 |
PDBsum entry 1ahh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ahh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.159
- 7alpha-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cholate + NAD+ = 3alpha,12alpha-dihydroxy-7-oxo-5beta-cholanate + NADH + H+
|
 |
 |
 |
 |
 |
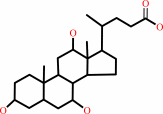 cholate
cholate
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
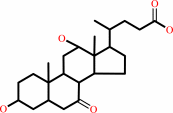 3alpha,12alpha-dihydroxy-7-oxo-5beta-cholanate
3alpha,12alpha-dihydroxy-7-oxo-5beta-cholanate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
35:7715-7730
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
|
|
N.Tanaka,
T.Nonaka,
T.Tanabe,
T.Yoshimoto,
D.Tsuru,
Y.Mitsui.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
7 alpha-Hydroxysteroid dehydrogenase (7 alpha-HSDH;1 EC 1.1.1.159) is an
NAD+-dependent oxidoreductase belonging to the short-chain
dehydrogenase/reductase (SDR) 1 family. It catalyzes the dehydrogenation of a
hydroxyl group at position 7 of the steroid skeleton of bile acids. The crystal
structure of the binary (complexed with NAD+) complex of 7 alpha-HSDH has been
solved at 2.3 A resolution by the multiple isomorphous replacement method. The
structure of the ternary complex [the enzyme complexed with NADH,
7-oxoglycochenodeoxycholic acid (as a reaction product), and possibly partially
glycochenodeoxycholic acid (as a substrate)] has been determined by a difference
Fourier method at 1.8 A resolution. The enzyme 7 alpha-HSDH is an alpha/beta
doubly wound protein having a Rossmann-fold domain for NAD (H) binding. Upon
substrate binding, large conformation changes occur at the substrate binding
loop (between the beta F strand and alpha G helix) and the C-terminal segment
(residues 250-255). The variable amino acid sequences of the substrate-binding
loop appear to be responsible for the wide variety of substrate specificities
observed among the enzymes of the SDR family. The crystal structure of the
ternary complex of 7 alpha-HSDH, which is the only structure available as the
ternary complex among the enzymes of the SDR family, indicates that the highly
conserved Tyr159 and Ser146 residues most probably directly interact with the
hydroxyl group of the substrates although this observation cannot be definite
due to an insufficiently characterized nature of the ternary complex. The
strictly conserved Lys163 is hydrogen-bonded to both the 2'- and 3'-hydroxyl
groups of the nicotinamide ribose of NAD(H). We propose a new catalytic
mechanism possibly common to all the enzymes belonging to the SDR family in
which a tyrosine residue (Tyr159) acts as a catalytic base and a serine residue
(Ser146) plays a subsidiary role of stabilizing substrate binding.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Katzberg,
N.Skorupa-Parachin,
M.F.Gorwa-Grauslund,
and
M.Bertau
(2010).
Engineering Cofactor Preference of Ketone Reducing Biocatalysts: A Mutagenesis Study on a gamma-Diketone Reductase from the Yeast Saccharomyces cerevisiae Serving as an Example.
|
| |
Int J Mol Sci,
11,
1735-1758.
|
 |
|
|
|
|
 |
K.Nakashima,
K.Ito,
Y.Nakajima,
R.Yamazawa,
S.Miyakawa,
and
T.Yoshimoto
(2009).
Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
|
| |
J Biochem,
145,
467-479.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Zhang,
H.Peng,
F.Gao,
Y.Liu,
H.Cheng,
J.Thompson,
and
G.F.Gao
(2009).
Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: Crystal structures of the substrate-free and quaternary complex enzymes.
|
| |
Protein Sci,
18,
294-303.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Miura,
T.Nishinaka,
and
T.Terada
(2008).
Different functions between human monomeric carbonyl reductase 3 and carbonyl reductase 1.
|
| |
Mol Cell Biochem,
315,
113-121.
|
 |
|
|
|
|
 |
T.P.Korman,
Y.H.Tan,
J.Wong,
R.Luo,
and
S.C.Tsai
(2008).
Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase.
|
| |
Biochemistry,
47,
1837-1847.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Zhou,
P.J.Enyeart,
and
C.O.Wilke
(2008).
Detecting clusters of mutations.
|
| |
PLoS ONE,
3,
e3765.
|
 |
|
|
|
|
 |
K.Palmu,
K.Ishida,
P.Mäntsälä,
C.Hertweck,
and
M.Metsä-Ketelä
(2007).
Artificial reconstruction of two cryptic angucycline antibiotic biosynthetic pathways.
|
| |
Chembiochem,
8,
1577-1584.
|
 |
|
|
|
|
 |
K.S.Paithankar,
C.Feller,
E.B.Kuettner,
A.Keim,
M.Grunow,
and
N.Sträter
(2007).
Cosubstrate-induced dynamics of D-3-hydroxybutyrate dehydrogenase from Pseudomonas putida.
|
| |
FEBS J,
274,
5767-5779.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.G.Nashev,
C.Chandsawangbhuwana,
Z.Balazs,
A.G.Atanasov,
B.Dick,
F.J.Frey,
M.E.Baker,
and
A.Odermatt
(2007).
Hexose-6-phosphate dehydrogenase modulates 11beta-hydroxysteroid dehydrogenase type 1-dependent metabolism of 7-keto- and 7beta-hydroxy-neurosteroids.
|
| |
PLoS ONE,
2,
e561.
|
 |
|
|
|
|
 |
O.B.Belyaeva,
and
F.F.Litvin
(2007).
Photoactive pigment-enzyme complexes of chlorophyll precursor in plant leaves.
|
| |
Biochemistry (Mosc),
72,
1458-1477.
|
 |
|
|
|
|
 |
H.Cho,
L.Huang,
A.Hamza,
D.Gao,
C.G.Zhan,
and
H.H.Tai
(2006).
Role of glutamine 148 of human 15-hydroxyprostaglandin dehydrogenase in catalytic oxidation of prostaglandin E2.
|
| |
Bioorg Med Chem,
14,
6486-6491.
|
 |
|
|
|
|
 |
J.A.Sundlov,
J.A.Garringer,
J.M.Carney,
A.S.Reger,
E.J.Drake,
W.L.Duax,
and
A.M.Gulick
(2006).
Determination of the crystal structure of EntA, a 2,3-dihydro-2,3-dihydroxybenzoic acid dehydrogenase from Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
734-740.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Aoki,
N.Tanaka,
S.Ishikura,
N.Araki,
Y.Imamura,
A.Hara,
and
K.T.Nakamura
(2006).
Crystallization and preliminary X-ray crystallographic studies of pig heart carbonyl reductase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1037-1040.
|
 |
|
|
|
|
 |
N.P.Ulrih,
and
T.Lanisnik Rizner
(2006).
Conformational stability of 17 beta-hydroxysteroid dehydrogenase from the fungus Cochliobolus lunatus.
|
| |
FEBS J,
273,
3927-3937.
|
 |
|
|
|
|
 |
S.Nakamura,
M.Oda,
S.Kataoka,
S.Ueda,
S.Uchiyama,
T.Yoshida,
Y.Kobayashi,
and
T.Ohkubo
(2006).
Apo- and holo-structures of 3alpha-hydroxysteroid dehydrogenase from Pseudomonas sp. B-0831. Loop-helix transition induced by coenzyme binding.
|
| |
J Biol Chem,
281,
31876-31884.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.G.Atanasov,
L.G.Nashev,
S.Tam,
M.E.Baker,
and
A.Odermatt
(2005).
Organotins disrupt the 11beta-hydroxysteroid dehydrogenase type 2-dependent local inactivation of glucocorticoids.
|
| |
Environ Health Perspect,
113,
1600-1606.
|
 |
|
|
|
|
 |
A.Philippsen,
T.Schirmer,
M.A.Stein,
F.Giffhorn,
and
J.Stetefeld
(2005).
Structure of zinc-independent sorbitol dehydrogenase from Rhodobacter sphaeroides at 2.4 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
374-379.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.J.Hosfield,
Y.Wu,
R.J.Skene,
M.Hilgers,
A.Jennings,
G.P.Snell,
and
K.Aertgeerts
(2005).
Conformational flexibility in crystal structures of human 11beta-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation.
|
| |
J Biol Chem,
280,
4639-4648.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Ogg,
B.Elleby,
C.Norström,
K.Stefansson,
L.Abrahmsén,
U.Oppermann,
and
S.Svensson
(2005).
The crystal structure of guinea pig 11beta-hydroxysteroid dehydrogenase type 1 provides a model for enzyme-lipid bilayer interactions.
|
| |
J Biol Chem,
280,
3789-3794.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.S.Alphey,
W.Yu,
E.Byres,
D.Li,
and
W.N.Hunter
(2005).
Structure and reactivity of human mitochondrial 2,4-dienoyl-CoA reductase: enzyme-ligand interactions in a distinctive short-chain reductase active site.
|
| |
J Biol Chem,
280,
3068-3077.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.El-Kabbani,
S.Ishikura,
C.Darmanin,
V.Carbone,
R.P.Chung,
N.Usami,
and
A.Hara
(2004).
Crystal structure of human L-xylulose reductase holoenzyme: probing the role of Asn107 with site-directed mutagenesis.
|
| |
Proteins,
55,
724-732.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Ueda,
M.Oda,
S.Imamura,
and
M.Ohnishi
(2004).
Transient-phase kinetic studies on the nucleotide binding to 3alpha-hydroxysteroid dehydrogenase from Pseudomonas sp. B-0831 using fluorescence stopped-flow procedures.
|
| |
Eur J Biochem,
271,
1774-1780.
|
 |
|
|
|
|
 |
A.Koumanov,
J.Benach,
S.Atrian,
R.Gonzàlez-Duarte,
A.Karshikoff,
and
R.Ladenstein
(2003).
The catalytic mechanism of Drosophila alcohol dehydrogenase: evidence for a proton relay modulated by the coupled ionization of the active site Lysine/Tyrosine pair and a NAD+ ribose OH switch.
|
| |
Proteins,
51,
289-298.
|
 |
|
|
|
|
 |
R.M.de Jong,
J.J.Tiesinga,
H.J.Rozeboom,
K.H.Kalk,
L.Tang,
D.B.Janssen,
and
B.W.Dijkstra
(2003).
Structure and mechanism of a bacterial haloalcohol dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site.
|
| |
EMBO J,
22,
4933-4944.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Lobedanz,
and
L.Søgaard-Andersen
(2003).
Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus.
|
| |
Genes Dev,
17,
2151-2161.
|
 |
|
|
|
|
 |
S.Sogabe,
A.Yoshizumi,
T.A.Fukami,
Y.Shiratori,
S.Shimizu,
H.Takagi,
S.Nakamori,
and
M.Wada
(2003).
The crystal structure and stereospecificity of levodione reductase from Corynebacterium aquaticum M-13.
|
| |
J Biol Chem,
278,
19387-19395.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.L.Duax,
V.Pletnev,
A.Addlagatta,
J.Bruenn,
and
C.M.Weeks
(2003).
Rational proteomics I. Fingerprint identification and cofactor specificity in the short-chain oxidoreductase (SCOR) enzyme family.
|
| |
Proteins,
53,
931-943.
|
 |
|
|
|
|
 |
Y.Yasutake,
S.Watanabe,
M.Yao,
Y.Takada,
N.Fukunaga,
and
I.Tanaka
(2003).
Crystal structure of the monomeric isocitrate dehydrogenase in the presence of NADP+: insight into the cofactor recognition, catalysis, and evolution.
|
| |
J Biol Chem,
278,
36897-36904.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Filling,
K.D.Berndt,
J.Benach,
S.Knapp,
T.Prozorovski,
E.Nordling,
R.Ladenstein,
H.Jörnvall,
and
U.Oppermann
(2002).
Critical residues for structure and catalysis in short-chain dehydrogenases/reductases.
|
| |
J Biol Chem,
277,
25677-25684.
|
 |
|
|
|
|
 |
F.Haeseleer,
G.F.Jang,
Y.Imanishi,
C.A.Driessen,
M.Matsumura,
P.S.Nelson,
and
K.Palczewski
(2002).
Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina.
|
| |
J Biol Chem,
277,
45537-45546.
|
 |
|
|
|
|
 |
B.Gerratana,
W.W.Cleland,
and
P.A.Frey
(2001).
Mechanistic roles of Thr134, Tyr160, and Lys 164 in the reaction catalyzed by dTDP-glucose 4,6-dehydratase.
|
| |
Biochemistry,
40,
9187-9195.
|
 |
|
|
|
|
 |
E.T.Johnson,
S.Ryu,
H.Yi,
B.Shin,
H.Cheong,
and
G.Choi
(2001).
Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase.
|
| |
Plant J,
25,
325-333.
|
 |
|
|
|
|
 |
H.Zhou,
F.Yan,
and
H.H.Tai
(2001).
C-Terminal region of human NAD+-dependent 15-hydroxyprostaglandin dehydrogenase is involved in the interaction with prostaglandin substrates.
|
| |
Eur J Biochem,
268,
3368-3374.
|
 |
|
|
|
|
 |
J.E.van Hylckama Vlieg,
L.Tang,
J.H.Lutje Spelberg,
T.Smilda,
G.J.Poelarends,
T.Bosma,
A.E.van Merode,
M.W.Fraaije,
and
D.B.Janssen
(2001).
Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases.
|
| |
J Bacteriol,
183,
5058-5066.
|
 |
|
|
|
|
 |
K.Fujimoto,
M.Hara,
H.Yamada,
M.Sakurai,
A.Inaba,
A.Tomomura,
and
S.Katoh
(2001).
Role of the conserved Ser-Tyr-Lys triad of the SDR family in sepiapterin reductase.
|
| |
Chem Biol Interact,
130,
825-832.
|
 |
|
|
|
|
 |
M.Otagiri,
G.Kurisu,
S.Swaminathan,
S.Ui,
S.Yoneda,
M.Ohkuma,
T.Kudo,
and
M.Kusunoki
(2001).
Crystallization and preliminary X-ray studies of meso-2,3-butanediol dehydrogenase from Klebsiella pneumoniae IAM1063.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
857-859.
|
 |
|
|
|
|
 |
S.Ishikura,
N.Usami,
K.Kitahara,
T.Isaji,
K.Oda,
J.Nakagawa,
and
A.Hara
(2001).
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family protein, P26h, in hamster testis and epididymis.
|
| |
Biochemistry,
40,
214-224.
|
 |
|
|
|
|
 |
T.Lanisnik Rizner,
J.Stojan,
and
J.Adamski
(2001).
17beta-hydroxysteroid dehydrogenase from the fungus Cochliobolus lunatus: structural and functional aspects.
|
| |
Chem Biol Interact,
130,
793-803.
|
 |
|
|
|
|
 |
D.A.Pelletier,
and
C.S.Harwood
(2000).
2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris.
|
| |
J Bacteriol,
182,
2753-2760.
|
 |
|
|
|
|
 |
J.Benach,
S.Atrian,
J.Fibla,
R.Gonzàlez-Duarte,
and
R.Ladenstein
(2000).
Structure-function relationships in Drosophila melanogaster alcohol dehydrogenase allozymes ADH(S), ADH(F) and ADH(UF), and distantly related forms.
|
| |
Eur J Biochem,
267,
3613-3622.
|
 |
|
|
|
|
 |
J.E.van Hylckama Vlieg,
H.Leemhuis,
J.H.Spelberg,
and
D.B.Janssen
(2000).
Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45.
|
| |
J Bacteriol,
182,
1956-1963.
|
 |
|
|
|
|
 |
M.Vedadi,
D.Barriault,
M.Sylvestre,
and
J.Powlowski
(2000).
Active site residues of cis-2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase from Comamonas testosteroni strain B-356.
|
| |
Biochemistry,
39,
5028-5034.
|
 |
|
|
|
|
 |
A.Yamashita,
H.Kato,
S.Wakatsuki,
T.Tomizaki,
T.Nakatsu,
K.Nakajima,
T.Hashimoto,
Y.Yamada,
and
J.Oda
(1999).
Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9 A resolution: implication for stereospecific substrate binding and catalysis.
|
| |
Biochemistry,
38,
7630-7637.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.A.Rozwarski,
C.Vilchèze,
M.Sugantino,
R.Bittman,
and
J.C.Sacchettini
(1999).
Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate.
|
| |
J Biol Chem,
274,
15582-15589.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.G.Gourley,
J.Luba,
L.W.Hardy,
S.M.Beverley,
and
W.N.Hunter
(1999).
Crystallization of recombinant Leishmania major pteridine reductase 1 (PTR1).
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
1608-1610.
|
 |
|
|
|
|
 |
J.N.Tinguely,
and
B.Wermuth
(1999).
Identification of the reactive cysteine residue (Cys227) in human carbonyl reductase.
|
| |
Eur J Biochem,
260,
9.
|
 |
|
|
|
|
 |
J.R.Allen,
and
S.A.Ensign
(1999).
Two short-chain dehydrogenases confer stereoselectivity for enantiomers of epoxypropane in the multiprotein epoxide carboxylating systems of Xanthobacter strain Py2 and Nocardia corallina B276.
|
| |
Biochemistry,
38,
247-256.
|
 |
|
|
|
|
 |
M.E.Baker
(1999).
TIP30, a cofactor for HIV-1 Tat-activated transcription, is homologous to short-chain dehydrogenases/reductases.
|
| |
Curr Biol,
9,
R471.
|
 |
|
|
|
|
 |
M.J.van der Werf,
C.van der Ven,
F.Barbirato,
M.H.Eppink,
J.A.de Bont,
and
W.J.van Berkel
(1999).
Stereoselective carveol dehydrogenase from Rhodococcus erythropolis DCL14. A novel nicotinoprotein belonging to the short chain dehydrogenase/reductase superfamily.
|
| |
J Biol Chem,
274,
26296-26304.
|
 |
|
|
|
|
 |
S.Menon,
M.Stahl,
R.Kumar,
G.Y.Xu,
and
F.Sullivan
(1999).
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from Escherichia coli.
|
| |
J Biol Chem,
274,
26743-26750.
|
 |
|
|
|
|
 |
C.Mazza,
R.Breton,
D.Housset,
and
J.C.Fontecilla-Camps
(1998).
Unusual charge stabilization of NADP+ in 17beta-hydroxysteroid dehydrogenase.
|
| |
J Biol Chem,
273,
8145-8152.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Möbus,
and
E.Maser
(1998).
Molecular cloning, overexpression, and characterization of steroid-inducible 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. A novel member of the short-chain dehydrogenase/reductase superfamily.
|
| |
J Biol Chem,
273,
30888-30896.
|
 |
|
|
|
|
 |
J.B.Thoden,
and
H.M.Holden
(1998).
Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli.
|
| |
Biochemistry,
37,
11469-11477.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Luba,
B.Nare,
P.H.Liang,
K.S.Anderson,
S.M.Beverley,
and
L.W.Hardy
(1998).
Leishmania major pteridine reductase 1 belongs to the short chain dehydrogenase family: stereochemical and kinetic evidence.
|
| |
Biochemistry,
37,
4093-4104.
|
 |
|
|
|
|
 |
K.Nakajima,
A.Yamashita,
H.Akama,
T.Nakatsu,
H.Kato,
T.Hashimoto,
J.Oda,
and
Y.Yamada
(1998).
Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
|
| |
Proc Natl Acad Sci U S A,
95,
4876-4881.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Torroja,
D.Ortuño-Sahagún,
A.Ferrús,
B.Hämmerle,
and
J.A.Barbas
(1998).
scully, an essential gene of Drosophila, is homologous to mammalian mitochondrial type II L-3-hydroxyacyl-CoA dehydrogenase/amyloid-beta peptide-binding protein.
|
| |
J Cell Biol,
141,
1009-1017.
|
 |
|
|
|
|
 |
M.E.McGrath,
J.T.Palmer,
D.Brömme,
and
J.R.Somoza
(1998).
Crystal structure of human cathepsin S.
|
| |
Protein Sci,
7,
1294-1302.
|
 |
|
|
|
|
 |
M.Hülsmeyer,
H.J.Hecht,
K.Niefind,
B.Hofer,
L.D.Eltis,
K.N.Timmis,
and
D.Schomburg
(1998).
Crystal structure of cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from a PCB degrader at 2.0 A resolution.
|
| |
Protein Sci,
7,
1286-1293.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.D.Mesecar,
B.L.Stoddard,
and
D.E.Koshland
(1997).
Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
|
| |
Science,
277,
202-206.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Auerbach,
A.Herrmann,
M.Gütlich,
M.Fischer,
U.Jacob,
A.Bacher,
and
R.Huber
(1997).
The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
|
| |
EMBO J,
16,
7219-7230.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
A.D.Hegeman,
G.Wesenberg,
M.C.Chapeau,
P.A.Frey,
and
H.M.Holden
(1997).
Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli.
|
| |
Biochemistry,
36,
6294-6304.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Nakanishi,
K.Matsuura,
H.Kaibe,
N.Tanaka,
T.Nonaka,
Y.Mitsui,
and
A.Hara
(1997).
Switch of coenzyme specificity of mouse lung carbonyl reductase by substitution of threonine 38 with aspartic acid.
|
| |
J Biol Chem,
272,
2218-2222.
|
 |
|
|
|
|
 |
P.M.Kiefer,
C.E.Grimshaw,
and
J.M.Whiteley
(1997).
The comparative interaction of quinonoid (6R)-dihydrobiopterin and an alternative dihydropterin substrate with wild-type and mutant rat dihydropteridine reductases.
|
| |
Biochemistry,
36,
9438-9445.
|
 |
|
|
|
|
 |
Y.Liu,
J.B.Thoden,
J.Kim,
E.Berger,
A.M.Gulick,
F.J.Ruzicka,
H.M.Holden,
and
P.A.Frey
(1997).
Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
|
| |
Biochemistry,
36,
10675-10684.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
P.A.Frey,
and
H.M.Holden
(1996).
High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol.
|
| |
Protein Sci,
5,
2149-2161.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.L.Duax,
J.F.Griffin,
and
D.Ghosh
(1996).
The fascinating complexities of steroid-binding enzymes.
|
| |
Curr Opin Struct Biol,
6,
813-823.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

