Elowitz and Leibler (2000), The Repressilator
July 2006, model of the month by Dominic P. Tolle
Original model: BIOMD0000000012
One of the major goals of Systems Biology is the elucidation of the control logic which determines the behaviour of naturally occuring biological systems[1]. To this end, Systems Biologist often create mathematical models designed to mimic a carefully observed biological system. Traditionally, the modeller aquires data, creates his model and tests the model against the available data. In an interesting take on the conventional way of modelling, Elowitz and Leibler[2] built a mathematical model of transcription regulation describing a cyclic negative-feedback loop made up of three repressor genes and their promoters. They used this model to determine the important parameters of the system and predict the systems behaviour, paying particular attention to parameter values that would cause the system to enter an unstable state leading to oscillatory behaviour. Finally the authors artifically reconstructd the system in E. coli using standard molecular biological approaches. In effect, rather than observing a natural system and explaining it in mathematical terms, the authors create a mathematical model to aid construction of an artificial control circiut. The result is an oscillating network which does not occur in nature, which the authors termed the Repressilator (see also the Brusselator[3] and the Oregonator[4] (BIOMD0000000040)).
The authors created a simple mathematical model of transcription regulation. The mathematical model was composed of six molecular species: three mRNA concentrations and three corresponding repressor protein concentrations. Each species was involved in transcription, translation and degradation reactions. Six coupled first-order differential equations described the dynamic behaviour of the system. Using the model, the authors predicted what parameters the stability of the steady state would be dependent on. In particular, the authors used the model to determine how to induce stable oscillations. Parameters that would favour oscillations were strong promoters, strong repression of transcription, cooperativity of repressor binding and similar lifetimes of mRNA and Proteins. The actual synthetic biological system was constructed from natural components using molecular biological techniques. Two alterations two the natural components were made to bring the system in line with the parameter space which favours oscillations: strong but tightly repressable hybrid promoters, and carboxy-terminal tags for the repressor proteins thus targetting them for protease degradation. A compatible reporter plasmid expressing GFP was also inserted into the system.

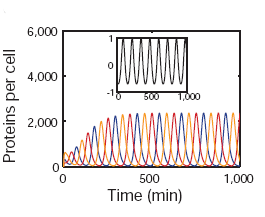
The temporal oscillations of fluorescence occured with a period of approximately 150 minutes, three times as long as the average cell-division time. Additionally, although sibling cells displayed correlated GFP levels for a long period of time after septation, there were variations in both period and amplitude.
Elowitz and Leibler are an excellent example of the use of mathematical models to design artificial control circuits. These can be either used as components for biotechnology and synthetic biology, or used to further study and understand properties of naturally evolved control elements.
Bibliographic References
- Kitano H (2002) Systems Biology: A Brief Overview. Science 295: 1662-1664.
- Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403: 335-338.
- P. Glansdorff and I. Prigogine, Thermodynamic theory of structure, stability and fluctuations. J. Wiley, Interscience, London, 1971
- Field RJ, Noyes RM (1974) Oscillations in chemical systems. IV. Limit cycle behavior in a model of a real chemical reaction. J. Chem. Phys. 60: 1877-1884
