Text Annotations (AI)
The text annotation tab gives AI-text mined residue-level functional annotations. A transformer-based deep learning model was fine-tuned to identify key terms in scientific literature that point to sequence and protein structure features. The found residues and point mutations mentioned in a publication text were linked to their corresponding residues in reference sequences from UniProt using the SIFTS resource. Furthermore, quality indicators for each found residue in a protein structure were collected from validation files and also linked to the text annotations.
Example entries used here:
- https://www.ebi.ac.uk/pdbe/entry/pdb/5cxt?activeTab=llm
- https://www.ebi.ac.uk/pdbe/entry/pdb/7zas?activeTab=llm
- Short video demonstrating page functionality on PDBe LinkedIn profile.
Layout of the page
The page has been organised into distinct sections. On the left, the macromolecules found in an entry are listed. This list can be collapsed to allow the vast central part containing the main information to become full-width.
In the centre, the layout contains five sections. At the top, we show a disclaimer regarding the fine-tuned model and where to find the annotations for download via our APIs. We also provide the details for the primary publication for the entry. Below, details about the source organism, UniProt reference sequence and the number of annotated residues are provided for a selected macromolecule. Following we have a sequence viewer. The last section is split into half with a table listing annotations on the left and Mol* viewer displaying the protein structure on the right. The sequence viewer, annotation table and Mol* viewer are all synchronised and respond to each other’s changes.
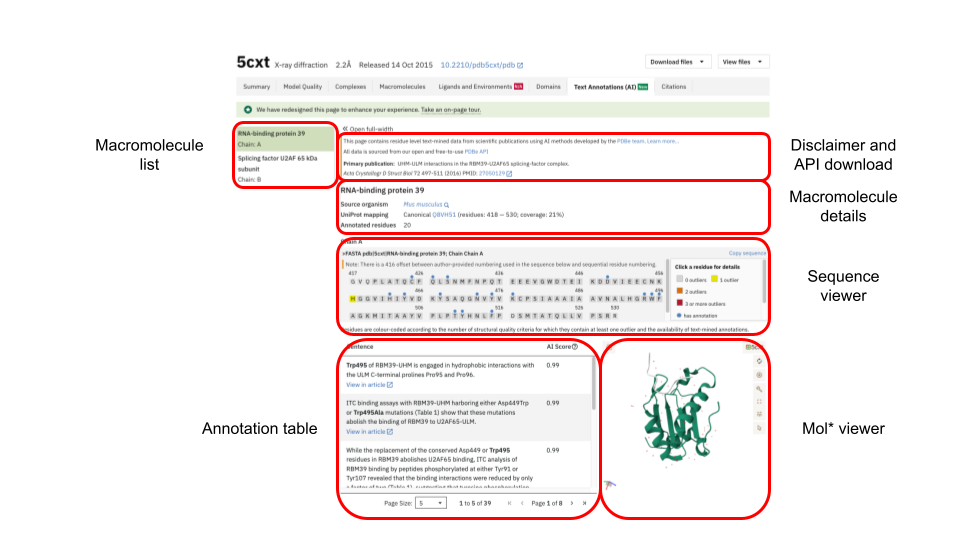
Macromolecule list
The macromolecule list displayed on the left is the same as one can find under the tab “Macromolecules”. It lists all macromolecules found in an entry, i.e. protein and nucleic acid. If a macromolecule’s chain in an entry has multiple copies then all the chain identifiers for the copies are given. Selecting a macromolecule from the list will update the sequence viewer, the annotation table and Mol* viewer for the selected entity.
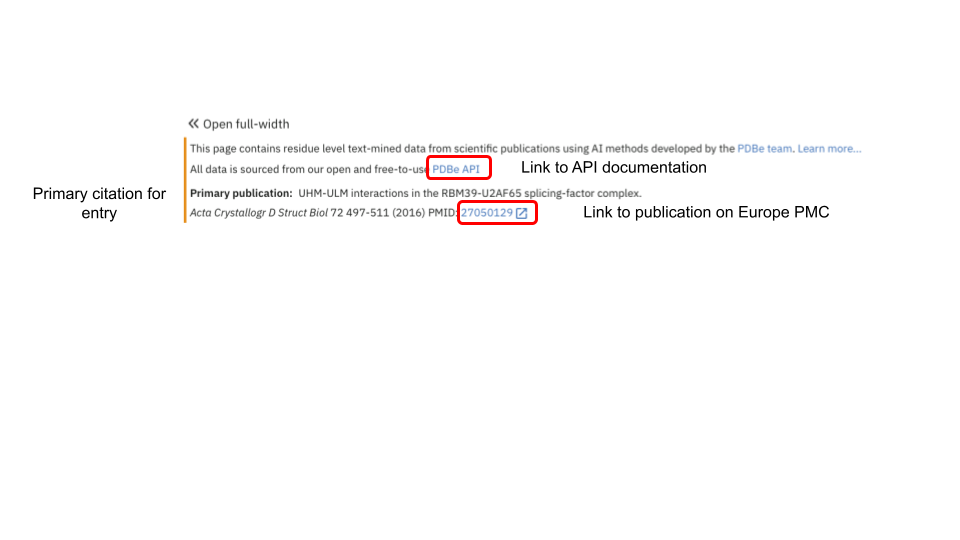
Disclaimer and API download
In this section, we provide details about the application programming interface (API) which underpins the information displayed on the webpage. Users can follow the link to the API documentation and explore the content through the interface or construct the required URLs for the download of annotations.
We also display the details for the primary citation of the entry, from which we also mined the annotations. If the publication is open access, then we provide a link to the document on Europe PMC via the article’s PubMed identifier.
Macromolecule details
Details about the selected macromolecule are given here. We display the source organism for the macromolecule and give its UniProt reference accession number. The covered residue span with respect to the reference sequence is given and how much this represents relative to the full sequence. Lastly, we give the number of residues for the selected macromolecule for which we have mined annotations

Sequence viewer
The sequence viewer is linked to the macromolecule list, the annotation table and Mol* viewer. Residues are highlighted for different properties:

Residues for which annotations have been mined are highlighted with a blue circle. Residues for which issues have been identified in the validation report are highlighted in a colour scheme, gray to dark red depending on severity of issues. Residues that were present in the purified sample but could not be modelled have gray, diagonal stripes.
Individual chains and residues can be selected from the sequence viewer and the corresponding chains, residues and annotations will be updated in Mol* viewer and the annotation table, respectively. Clicking on the same residue again will de-select it and restore viewers and the table to the same state as when first landing on the page.

A note is presented pointing to potential sequence offsets, if it applies. Offsets occur regularly when depositors select to start residue numbering of their structure with “1” instead of the corresponding sequence position from a UniProt reference sequence.
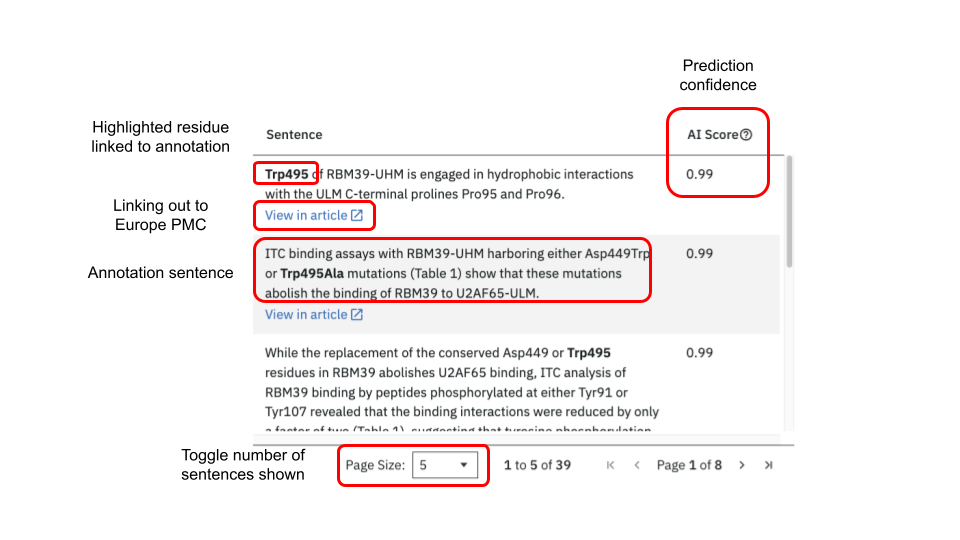
Annotation table
When first opening the annotation tab all annotations found for an entry will be in the central table. Upon selecting a macromolecule, chain and residue only the specific subset of annotations for the selected residue will be given. In each sentence the residue of interest is highlighted in bold. The highlighted residues in the sentence are themselves clickable and will update the sequence viewer and Mol* viewer in turn. For each identified residue a confidence score for the prediction is given as “AI Score”. If a publication is open access then clicking on “View in article” opens a new page to PubMed Central centering on and highlighting the sentence within the publication. The number of sentences displayed in the table can be set to 5, 10 or 20.
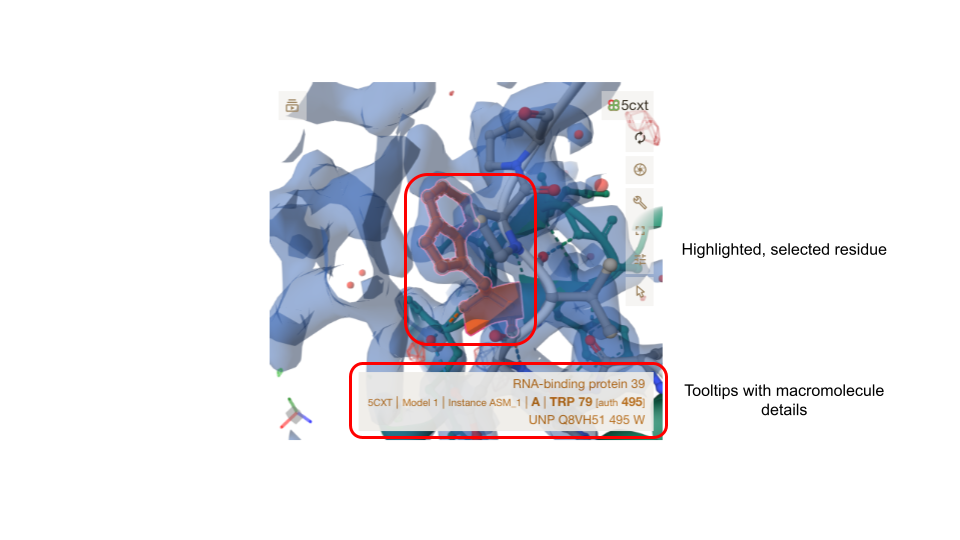
Mol* viewer
Any selected macromolecule of an entry has its atomic coordinates displayed in Mol* viewer. The viewer itself is well documented, and in the context of the residue-level annotation,s only its basic functionality is used. When first landing on the page, the whole molecule for the first macromolecule in an entry file is displayed in cartoon representation, coloured green. Selecting a residue from the sequence viewer, the annotation table or directly in Mol* will centre the molecule on this residue and display it in ball-and-stick representation, coloured orange. Surrounding residues of interest will appear as ball-and-stick representations, and interactions are indicated through dashed lines. Where experimental data is available, electron density maps or electric potential maps will be displayed. Hovering over a residue will bring a tool-tip box to the fore, giving details about the residue in the structure and its reference in a UniProt accession.
Missing annotations
A statement is displayed on the page if the primary citation linked to an entry could not be mined for text annotations (see below).
Residue-level annotations may be lacking due to issues such as absent or mismatched residue numbering in publication texts, restricted access to full-text articles, or missing user-submitted data. Additionally, the annotation pipeline might have failed to extract relevant information or accurately map it to structural data.
The lack of annotations can have a number of reasons:
- The primary citation is not open access, and we were not able to access the manuscript due to copyright issues.
- Our collaborators did not submit annotations to our system for processing and display.
- The predictive model was not able to identify any residue-level functional annotations.
- Found residues could not be validated or linked to a UniProt reference.