Proteomics resources at EMBL-EBI
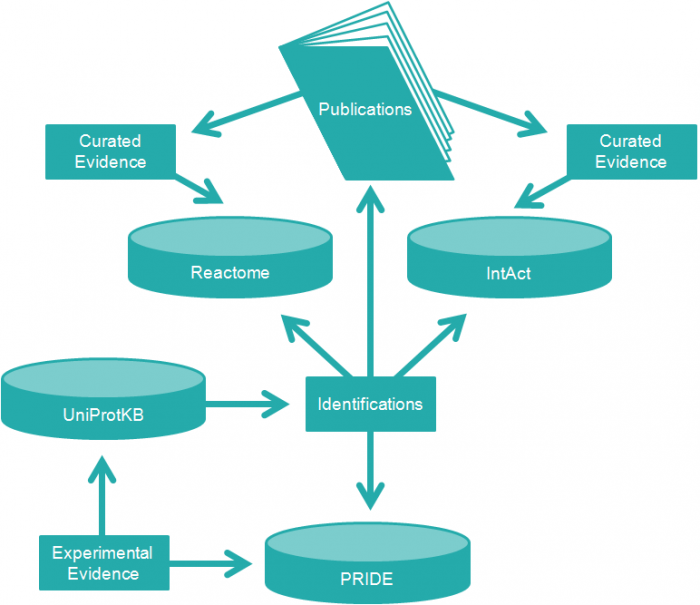
This section will introduce you to the proteomics resources at EMBL-EBI: UniProt, IntAct, Reactome and PRIDE (Figure 2):
- The UniProt Knowledgebase (UniProtKB) database contains protein sequences and information about the known biological functions (e.g ‘an ATP synthase’) of proteins (1)
- The IntAct database contains information about protein interactions (2)
- The Reactome database contains information about which roles proteins play in human biological pathways, and which processes they contribute to (3)
- The PRIDE database contains experimental evidence of published protein and peptide identifications (4)