Gene Ontology
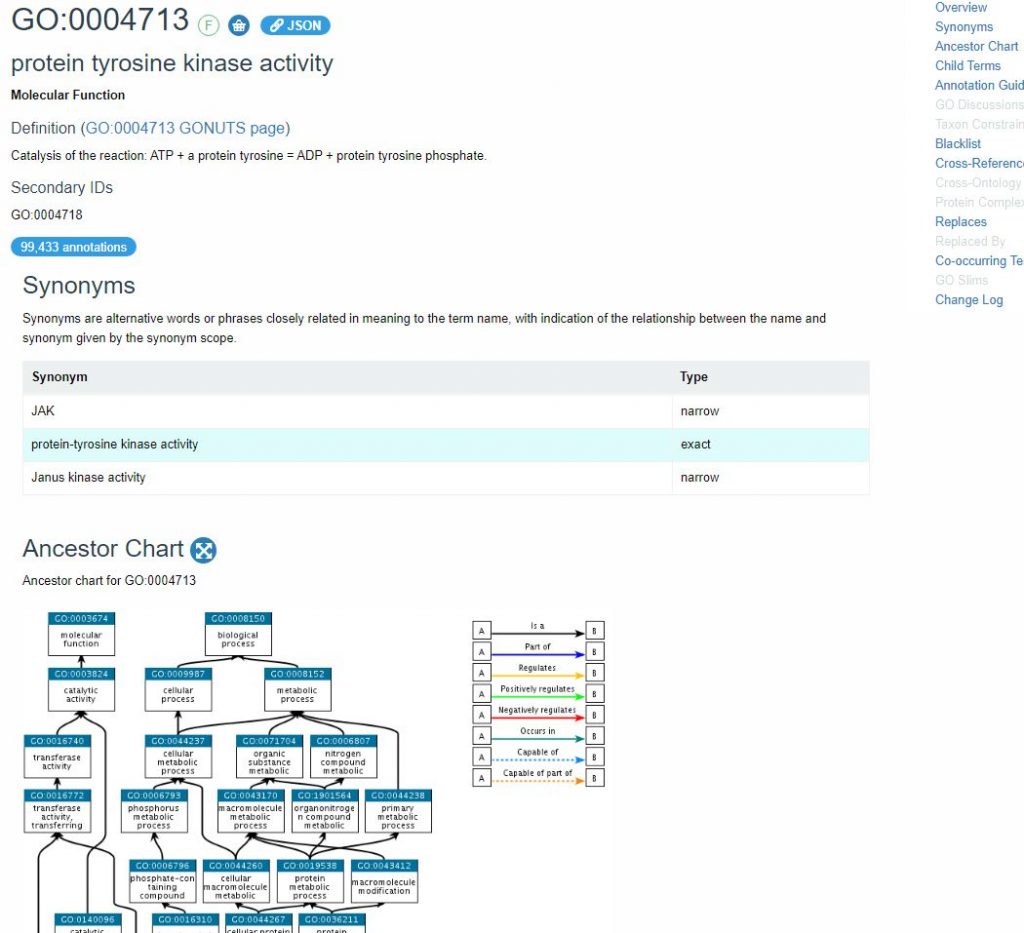
The archetypal example of an ontology in the molecular life sciences is the Gene Ontology (GO), created and maintained by the Gene Ontology Consortium. GO describes the function and cellular localisation of gene products across all species (Figure 9), and you can find out more about it in our GOA and QuickGo: Quick tour.
GO is used to describe genes and their products in major public databases, including UniProt, Ensembl and many model organism databases such as FlyBase, SGD and MGI. It provides a powerful means of analysing data sets, for example, it can be used to determine if genes identified as over-expressed in a functional genomics experiment have related functions or similar locations (GO enrichment analysis). For more on Gene Ontology, watch the recorded webinar Exploring Gene Ontology, GO annotations, tools and resources.
Since the development of GO in the late 1990s, many more ontologies have been developed using the same principles. These principles include open access, collaborative development and interoperability. These ontologies are collected in the OBO Foundry and are also listed at FAIRsharing.org.
If you have a data set and want to annotate it using an ontology, it can be quite a challenge to identify the most appropriate one. The Ontology Lookup Service is a good place to start, requiring no prior knowledge of what ontologies exist.