 |
PDBsum entry 7wpk
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Methionyl-tRNA synthetase from staphylococcus aureus complexed with l- met
|
|
Structure:
|
 |
Methionine--tRNA ligase. Chain: a. Synonym: methionyl-tRNA synthetase,metrs. Engineered: yes
|
|
Source:
|
 |
Staphylococcus aureus subsp. Aureus col. Staphylococcus aureus col. Organism_taxid: 93062. Strain: col. Gene: metg. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.232
|
R-free:
|
0.279
|
|
|
Authors:
|
 |
J.Yi,Z.Cai,H.Qiu,F.Lu,B.Chen,Z.Luo,Q.Gu,J.Xu,H.Zhou
|
|
Key ref:
|
 |
J.Yi
et al.
Fragment screening and structural analyses highlight ATP-Assisted ligand binding for inhibitor discovery a type 1 methionyl-Trna synthetase.
Nucleic acids res,
.
PubMed id:
|
 |
|
Date:
|
 |
|
24-Jan-22
|
Release date:
|
27-Apr-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5HII6
(SYM_STAAC) -
Methionine--tRNA ligase from Staphylococcus aureus (strain COL)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
657 a.a.
512 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.10
- methionine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Met) + L-methionine + ATP = L-methionyl-tRNA(Met) + AMP + diphosphate
|
 |
 |
 |
 |
 |
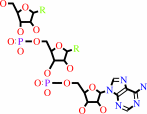 tRNA(Met)
tRNA(Met)
|
+
|
L-methionine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
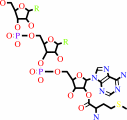 L-methionyl-tRNA(Met)
L-methionyl-tRNA(Met)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

