 |
PDBsum entry 6o1h
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Tryptophan synthase q114a mutant in complex with n-(4'- trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (f9f) at the enzyme alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid at the enzyme beta site
|
|
Structure:
|
 |
Tryptophan synthase alpha chain. Chain: a. Engineered: yes. Tryptophan synthase beta chain. Chain: b. Engineered: yes. Mutation: yes. Other_details: beta-chain contains mutation q114a
|
|
Source:
|
 |
Salmonella typhimurium. Salmonella enterica subsp. Enterica serovar typhimurium. Organism_taxid: 90371. Gene: trpa, dd95_04145. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008. Gene: trpb, stm1726.
|
|
Resolution:
|
 |
|
1.64Å
|
R-factor:
|
0.156
|
R-free:
|
0.199
|
|
|
Authors:
|
 |
E.Hilario,M.F.Dunn,L.J.Mueller,L.Fan
|
|
Key ref:
|
 |
E.Hilario
et al.
Crystal structure of salmonella typhimurium tryptopha synthase mutant beta-Q114a with 2-({[4-(Trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (f9f) at the alpha-Site, Cesium the metal coordination site, And [3-Hydroxy-2-Methyl-5-Phosphonooxymethyl-Pyridin-4-Yl -Serine (pls) at the beta-Site..
To be published,
.
|
 |
|
Date:
|
 |
|
19-Feb-19
|
Release date:
|
19-Feb-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
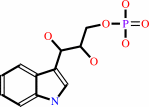 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 L-serine
L-serine
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
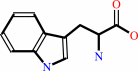 L-tryptophan
L-tryptophan
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 46.67% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
1D0)
matches with 50.00% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

