 |
PDBsum entry 5yas

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.47
- (S)-hydroxynitrile lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a monosubstituted aliphatic (S)-hydroxynitrile = an aldehyde + hydrogen cyanide
|
|
2.
|
a disubstituted aliphatic (S)-hydroxynitrile = a ketone + hydrogen cyanide
|
|
3.
|
an aromatic (S)-hydroxynitrile = an aromatic aldehyde + hydrogen cyanide
|
|
 |
 |
 |
 |
 |
monosubstituted aliphatic (S)-hydroxynitrile
|
=
|
 aldehyde
aldehyde
|
+
|
 hydrogen cyanide
hydrogen cyanide
|
|
 |
 |
 |
 |
 |
disubstituted aliphatic (S)-hydroxynitrile
|
=
|
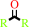 ketone
ketone
|
+
|
 hydrogen cyanide
hydrogen cyanide
|
|
 |
 |
 |
 |
 |
aromatic (S)-hydroxynitrile
|
=
|
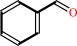 aromatic aldehyde
aromatic aldehyde
|
+
|
 hydrogen cyanide
hydrogen cyanide
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
8:1990-2000
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
|
|
J.Zuegg,
K.Gruber,
M.Gugganig,
U.G.Wagner,
C.Kratky.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The 3D structures of complexes between the hydroxynitrile lyase from Hevea
brasiliensis (Hb-HNL) and several substrate and/or inhibitor molecules,
including trichloracetaldehyde, hexafluoracetone, acetone, and rhodanide, were
determined by X-ray crystallography. The complex with trichloracetaldehyde
showed a covalent linkage between the protein and the inhibitor, which had
apparently resulted from nucleophilic attack of the catalytic Ser80-Ogamma. All
other complexes showed the substrate or inhibitor molecule merely hydrogen
bonded to the protein. In addition, the native crystal structure of Hb-HNL was
redetermined at cryo-temperature and at room temperature, eliminating previous
uncertainties concerning residual electron density within the active site, and
leading to the observation of two conserved water molecules. One of them was
found to be conserved in all complex structures and appears to have mainly
structural significance. The other water molecule is conserved in all structures
except for the complex with rhodanide; it is hydrogen bonded to the imidazole of
the catalytic His235 and appears to affect the Hb-HNL catalyzed reaction. The
observed 3D structural data suggest implications for the enzyme mechanism. It
appears that the enzyme-catalyzed cyanohydrin formation is unlikely to proceed
via a hemiacetal or hemiketal intermediate covalently attached to the enzyme,
despite the observation of such an intermediate for the complex with
trichloracetaldehyde. Instead, the data are consistent with a mechanism where
the incoming substrate is activated by hydrogen bonding with its carbonyl oxygen
to the Ser80 and Thr11 hydroxy groups. A hydrogen cyanide molecule subsequently
replaces a water molecule and is deprotonated presumably by the His235 base.
Deprotonation is facilitated by the proximity of the positive charge of the
Lys236 side chain.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 5.
Fig. 5. Residual density within the active site of Hb--HNL for the ~A! F6-acetone, ~B! rhodanide, and ~C! acetone soaks. See caption
to Figure 3 for details.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from the Protein Society:
Protein Sci
(1999,
8,
1990-2000)
copyright 1999.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Dreveny,
A.S.Andryushkova,
A.Glieder,
K.Gruber,
and
C.Kratky
(2009).
Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
|
| |
Biochemistry,
48,
3370-3377.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.C.Bas,
D.M.Rogers,
and
J.H.Jensen
(2008).
Very fast prediction and rationalization of pKa values for protein-ligand complexes.
|
| |
Proteins,
73,
765-783.
|
 |
|
|
|
|
 |
M.Avi,
R.M.Wiedner,
H.Griengl,
and
H.Schwab
(2008).
Improvement of a stereoselective biocatalytic synthesis by substrate and enzyme engineering: 2-hydroxy-(4'-oxocyclohexyl)acetonitrile as the model.
|
| |
Chemistry,
14,
11415-11422.
|
 |
|
|
|
|
 |
Q.Luo,
W.W.Han,
Y.H.Zhou,
Y.Yao,
and
Z.S.Li
(2008).
The 3D structure of the defense-related rice protein Pir7b predicted by homology modeling and ligand binding studies.
|
| |
J Mol Model,
14,
559-569.
|
 |
|
|
|
|
 |
J.J.Li,
and
T.D.Bugg
(2007).
Investigation of a general base mechanism for ester hydrolysis in C-C hydrolase enzymes of the alpha/beta-hydrolase superfamily: a novel mechanism for the serine catalytic triad.
|
| |
Org Biomol Chem,
5,
507-513.
|
 |
|
|
|
|
 |
B.D.Charette,
R.G.Macdonald,
S.Wetzel,
D.B.Berkowitz,
and
H.Waldmann
(2006).
Protein structure similarity clustering: dynamic treatment of PDB structures facilitates clustering.
|
| |
Angew Chem Int Ed Engl,
45,
7766-7770.
|
 |
|
|
|
|
 |
T.Purkarthofer,
K.Gruber,
M.Gruber-Khadjawi,
K.Waich,
W.Skranc,
D.Mink,
and
H.Griengl
(2006).
A biocatalytic Henry reaction--the hydroxynitrile lyase from Hevea brasiliensis also catalyzes nitroaldol reactions.
|
| |
Angew Chem Int Ed Engl,
45,
3454-3456.
|
 |
|
|
|
|
 |
R.Xu,
M.H.Zong,
Y.Y.Liu,
J.He,
Y.Y.Zhang,
and
W.Y.Lou
(2004).
Enzymatic enantioselective transcyanation of silicon-containing aliphatic ketone with (S)-hydroxynitrile lyase from Manihot esculenta.
|
| |
Appl Microbiol Biotechnol,
66,
27-33.
|
 |
|
|
|
|
 |
H.Bühler,
F.Effenberger,
S.Förster,
J.Roos,
and
H.Wajant
(2003).
Substrate specificity of mutants of the hydroxynitrile lyase from Manihot esculenta.
|
| |
Chembiochem,
4,
211-216.
|
 |
|
|
|
|
 |
H.Lauble,
S.Förster,
B.Miehlich,
H.Wajant,
and
F.Effenberger
(2001).
Structure of hydroxynitrile lyase from Manihot esculenta in complex with substrates acetone and chloroacetone: implications for the mechanism of cyanogenesis.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
194-200.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.V.Johnson,
A.A.Zabelinskaja-Mackova,
and
H.Griengl
(2000).
Oxynitrilases for asymmetric C-C bond formation.
|
| |
Curr Opin Chem Biol,
4,
103-109.
|
 |
|
|
|
|
 |
F.Effenberger,
S.Förster,
and
H.Wajant
(2000).
Hydroxynitrile lyases in stereoselective catalysis.
|
| |
Curr Opin Biotechnol,
11,
532-539.
|
 |
|
|
|
|
 |
H.Griengl,
H.Schwab,
and
M.Fechter
(2000).
The synthesis of chiral cyanohydrins by oxynitrilases.
|
| |
Trends Biotechnol,
18,
252-256.
|
 |
|
|
|
|
 |
R.J.Kazlauskas
(2000).
Molecular modeling and biocatalysis: explanations, predictions, limitations, and opportunities.
|
| |
Curr Opin Chem Biol,
4,
81-88.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

