 |
PDBsum entry 4lrd
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.4.2.7
- phosphopentomutase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-ribose 1-phosphate = D-ribose 5-phosphate
|
 |
 |
 |
 |
 |
alpha-D-ribose 1-phosphate
Bound ligand (Het Group name = )
matches with 42.86% similarity
|
=
|
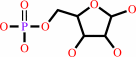 D-ribose 5-phosphate
D-ribose 5-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Chem Biol
10:392-399
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Bioretrosynthetic construction of a didanosine biosynthetic pathway.
|
|
W.R.Birmingham,
C.A.Starbird,
T.D.Panosian,
D.P.Nannemann,
T.M.Iverson,
B.O.Bachmann.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Concatenation of engineered biocatalysts into multistep pathways markedly
increases their utility, but the development of generalizable assembly methods
remains a major challenge. Herein we evaluate 'bioretrosynthesis', which is an
application of the retrograde evolution hypothesis, for biosynthetic pathway
construction. To test bioretrosynthesis, we engineered a pathway for synthesis
of the antiretroviral nucleoside analog didanosine (2',3'-dideoxyinosine).
Applying both directed evolution- and structure-based approaches, we began
pathway construction with a retro-extension from an engineered purine nucleoside
phosphorylase and evolved 1,5-phosphopentomutase to accept the substrate
2,3-dideoxyribose 5-phosphate with a 700-fold change in substrate selectivity
and threefold increased turnover in cell lysate. A subsequent retrograde pathway
extension, via ribokinase engineering, resulted in a didanosine pathway with a
9,500-fold change in nucleoside production selectivity and 50-fold increase in
didanosine production. Unexpectedly, the result of this bioretrosynthetic step
was not a retro-extension from phosphopentomutase but rather the discovery of a
fortuitous pathway-shortening bypass via the engineered ribokinase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

