 |
PDBsum entry 4jzr

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4jzr
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Structure of prolyl hydroxylase domain-containing protein (phd) with inhibitors
|
|
Structure:
|
 |
Egl nine homolog 1. Chain: a. Fragment: unp residues 189-399. Synonym: hypoxia-inducible factor prolyl hydroxylase 2, hif-ph2, hif- prolyl hydroxylase 2, hph-2, prolyl hydroxylase domain-containing protein 2, phd2, sm-20. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: c1orf12, egln1, pnas-118, pnas-137. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.232
|
R-free:
|
0.259
|
|
|
Authors:
|
 |
Y.Ma,L.Yang
|
|
Key ref:
|
 |
G.Deng
et al.
(2013).
Novel complex crystal structure of prolyl hydroxylase domain-containing protein 2 (PHD2): 2,8-Diazaspiro[4.5]decan-1-ones as potent, orally bioavailable PHD2 inhibitors.
Bioorg Med Chem Lett,
21,
6349-6358.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
03-Apr-13
|
Release date:
|
30-Oct-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9GZT9
(EGLN1_HUMAN) -
Egl nine homolog 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
426 a.a.
204 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.11.29
- hypoxia-inducible factor-proline dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-prolyl-[hypoxia-inducible factor alpha subunit] + 2-oxoglutarate + O2 = trans-4-hydroxy-L-prolyl-[hypoxia-inducible factor alpha subunit] + succinate + CO2
|
 |
 |
 |
 |
 |
L-prolyl-[hypoxia-inducible factor alpha subunit]
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
trans-4-hydroxy-L-prolyl-[hypoxia-inducible factor alpha subunit]
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+); L-ascorbate
|
 |
 |
 |
 |
 |
Fe(2+)
|
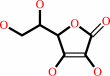 L-ascorbate
L-ascorbate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Bioorg Med Chem Lett
21:6349-6358
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Novel complex crystal structure of prolyl hydroxylase domain-containing protein 2 (PHD2): 2,8-Diazaspiro[4.5]decan-1-ones as potent, orally bioavailable PHD2 inhibitors.
|
|
G.Deng,
B.Zhao,
Y.Ma,
Q.Xu,
H.Wang,
L.Yang,
Q.Zhang,
T.B.Guo,
W.Zhang,
Y.Jiao,
X.Cai,
J.Zhang,
H.Liu,
X.Guan,
H.Lu,
J.Xiang,
J.D.Elliott,
X.Lin,
F.Ren.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We have discovered a novel complex crystal structure of the PHD2 enzyme with its
inhibitor, the 2,8-diazaspiro[4.5]decan-1-one analogue 4b. The widely reported
salt bridge between Arg383 of the enzyme and its inhibitors in all complex
structures published thus far was not observed in our case. In our complex
structure compound 4b forms several novel interactions with the enzyme, which
include a hydrogen bond with Arg322, a π-cation interaction with Arg322, a
π-π stacking with Trp389, and a π-π stacking with His313. Guided by the
structural information, SAR studies were performed on the
2,8-diazaspiro[4.5]decan-1-one series leading to the discovery of compound 9p
with high potency and good oral pharmacokinetic profile in mice.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

