 |
PDBsum entry 4c5c
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
The x-ray crystal structure of d-alanyl-d-alanine ligase in complex with adp and d-ala-d-ala
|
|
Structure:
|
 |
D-alanine--d-alanine ligase. Chain: a, b. Synonym: d-ala-d-ala ligase, d-alanylalanine synthetase, d-alanyl-d- alanine ligase. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 83333. Strain: k-12. Expressed in: escherichia coli. Expression_system_taxid: 469008. Expression_system_variant: rosetta.
|
|
Resolution:
|
 |
|
1.40Å
|
R-factor:
|
0.181
|
R-free:
|
0.203
|
|
|
Authors:
|
 |
S.Batson,V.Majce,A.J.Lloyd,D.Rea,C.W.G.Fishwick,K.J.Simmons,V.Fulop, D.I.Roper
|
|
Key ref:
|
 |
S.Batson
et al.
(2017).
Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun,
8,
1939.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Sep-13
|
Release date:
|
21-Jan-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07862
(DDLB_ECOLI) -
D-alanine--D-alanine ligase B from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
306 a.a.
306 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.4
- D-alanine--D-alanine ligase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Peptidoglycan Biosynthesis (Part 1)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 D-alanine + ATP = D-alanyl-D-alanine + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
2
×
D-alanine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
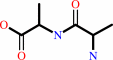 D-alanyl-D-alanine
D-alanyl-D-alanine
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Commun
8:1939
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
|
|
S.Batson,
C.de Chiara,
V.Majce,
A.J.Lloyd,
S.Gobec,
D.Rea,
V.Fülöp,
C.W.Thoroughgood,
K.J.Simmons,
C.G.Dowson,
C.W.G.Fishwick,
L.P.S.de Carvalho,
D.I.Roper.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
D-cycloserine is an antibiotic which targets sequential bacterial cell wall
peptidoglycan biosynthesis enzymes: alanine racemase and D-alanine:D-alanine
ligase. By a combination of structural, chemical and mechanistic studies here we
show that the inhibition of D-alanine:D-alanine ligase by the antibiotic
D-cycloserine proceeds via a distinct phosphorylated form of the drug. This
mechanistic insight reveals a bimodal mechanism of action for a single
antibiotic on different enzyme targets and has significance for the design of
future inhibitor molecules based on this chemical structure.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

