 |
PDBsum entry 4b4x
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of a complex between actinomadura r39 dd-peptidase and a sulfonamide boronate inhibitor
|
|
Structure:
|
 |
D-alanyl-d-alanine carboxypeptidase. Chain: a, b, c, d. Fragment: residues 50-515. Synonym: dd-carboxypeptidase, dd-peptidase, penicillin-binding protein, pbp. Ec: 3.4.16.4
|
|
Source:
|
 |
Actinomadura sp r39. Organism_taxid: 72570
|
|
Resolution:
|
 |
|
2.65Å
|
R-factor:
|
0.220
|
R-free:
|
0.268
|
|
|
Authors:
|
 |
S.E.Cannella,E.Sauvage,R.Herman,F.Kerff,M.Rocaboy,P.Charlier
|
|
Key ref:
|
 |
E.Sauvage
Crystal structure of a complex between actinomadura r dd-Peptidase and a boronate inhibitor.
To be published,
.
|
 |
|
Date:
|
 |
|
01-Aug-12
|
Release date:
|
21-Aug-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P39045
(DAC_ACTSP) -
D-alanyl-D-alanine carboxypeptidase from Actinomadura sp. (strain R39)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
538 a.a.
466 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.16.4
- serine-type D-Ala-D-Ala carboxypeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-alanyl-D-alanine + H2O = 2 D-alanine
|
 |
 |
 |
 |
 |
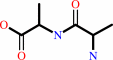
|
+
|
|
=
|
 2
×
2
×
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

