 |
PDBsum entry 3vmm
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.49
- L-alanine--L-anticapsin ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-anticapsin + L-alanine + ATP = bacilysin + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
L-anticapsin
|
+
|
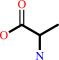 L-alanine
L-alanine
|
+
|
 ATP
ATP
|
=
|
bacilysin
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+) or Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Protein Sci
21:707-716
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and enzymatic characterization of BacD, an L-amino acid dipeptide ligase from Bacillus subtilis.
|
|
Y.Shomura,
E.Hinokuchi,
H.Ikeda,
A.Senoo,
Y.Takahashi,
J.Saito,
H.Komori,
N.Shibata,
Y.Yonetani,
Y.Higuchi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

