 |
PDBsum entry 3nxk

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 1 more)
327 a.a.
(+ 1 more)
327 a.a.
|
 |

|

|

|

|

|

|

|
 306 a.a.
306 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of probable cytoplasmic l-asparaginase from campylobacter jejuni
|
|
Structure:
|
 |
Cytoplasmic l-asparaginase. Chain: a, b, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Campylobacter jejuni subsp. Jejuni. Organism_taxid: 192222. Strain: nctc 11168. Gene: ansa, cj0029. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.169
|
R-free:
|
0.228
|
|
|
Authors:
|
 |
Y.Kim,M.Makowska-Grzyska,N.Maltseva,L.Papazisi,W.F.Anderson, A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
Y.Kim
et al.
Crystal structure of probable cytoplasmic l-Asparagin from campylobacter jejuni.
To be published,
.
|
 |
|
Date:
|
 |
|
14-Jul-10
|
Release date:
|
04-Aug-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.3.5.1.1
- asparaginase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-asparagine + H2O = L-aspartate + NH4+
|
 |
 |
 |
 |
 |
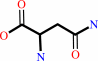 L-asparagine
L-asparagine
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 44.44% similarity
|
=
|
 L-aspartate
L-aspartate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

