 |
PDBsum entry 3grf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.3
- ornithine carbamoyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Urea Cycle and Arginine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
carbamoyl phosphate + L-ornithine = L-citrulline + phosphate + H+
|
 |
 |
 |
 |
 |
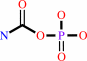 carbamoyl phosphate
carbamoyl phosphate
|
+
|
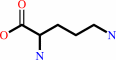 L-ornithine
L-ornithine
|
=
|
 L-citrulline
L-citrulline
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
76:1049-1053
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray structure and kinetic properties of ornithine transcarbamoylase from the human parasite Giardia lamblia.
|
|
A.Galkin,
L.Kulakova,
R.Wu,
M.Gong,
D.Dunaway-Mariano,
O.Herzberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. Overall fold, active site architecture of glOTC, and
sequence alignment of glOTC and hOTC. (A) Ribbon diagram
representation of the protein trimer. The Ni^2+ ion at the
center of the trimer is shown as magenta sphere. (B)
Stereoscopic view of the glOTC active site superposed with the
hOTC/PALO active site. The carbon atoms are colored green
(glOTC) and magenta (hOTC). Other atomic colors are as follows:
oxygen, red; nitrogen, blue; phosphor, orange; and sulfur,
yellow. Black residue labels correspond to glOTC, except that
the two residues labeled in magenta color (His117 of a
neighboring subunit and Met268) are hOTC residues that are
disordered in the glOTC structure (Ser82 and Tyr245,
respectively). (C) Sequence alignment of glOTC and hOTC.
Identical residues are blocked in blue and residues surrounding
the active site are indicated by red triangles.
|
 |
|
|

|
| |
The above figure is
reprinted
from an Open Access publication published by John Wiley & Sons, Inc.:
Proteins
(2009,
76,
1049-1053)
copyright 2009.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Galkin,
L.Kulakova,
R.Wu,
T.E.Nash,
D.Dunaway-Mariano,
and
O.Herzberg
(2010).
X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
386-390.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

