 |
PDBsum entry 3f0t

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with n-methyl-dhbt
|
|
Structure:
|
 |
Thymidine kinase. Chain: a, b. Fragment: residues 45-376. Engineered: yes
|
|
Source:
|
 |
Herpes simplex virus (type 1 / strain 17). Virus. Organism_taxid: 10299. Strain: 17. Gene: tk, ul23. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.195
|
R-free:
|
0.228
|
|
|
Authors:
|
 |
L.Pernot,R.Perozzo,Y.Westermaier,M.Martic,S.Ametamey,L.Scapozza
|
|
Key ref:
|
 |
M.Martić
et al.
(2011).
Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids,
30,
293-315.
PubMed id:
|
 |
|
Date:
|
 |
|
26-Oct-08
|
Release date:
|
03-Nov-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0DTH5
(KITH_HHV11) -
Thymidine kinase from Human herpesvirus 1 (strain 17)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
376 a.a.
309 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.21
- thymidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
thymidine + ATP = dTMP + ADP + H+
|
 |
 |
 |
 |
 |
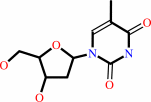 thymidine
thymidine
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 73.68% similarity
|
=
|
 dTMP
dTMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nucleosides Nucleotides Nucleic Acids
30:293-315
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
|
|
M.Martić,
L.Pernot,
Y.Westermaier,
R.Perozzo,
T.G.Kraljević,
S.Krištafor,
S.Raić-Malić,
L.Scapozza,
S.Ametamey.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

