 |
PDBsum entry 3dyd

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Human tyrosine aminotransferase
|
|
Structure:
|
 |
Tyrosine aminotransferase. Chain: a, b. Synonym: l-tyrosine:2-oxoglutarate aminotransferase, tat. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: tat. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.222
|
R-free:
|
0.261
|
|
|
Authors:
|
 |
T.Karlberg,M.Moche,J.Andersson,C.H.Arrowsmith,H.Berglund,R.Collins, L.G.Dahlgren,A.M.Edwards,S.Flodin,A.Flores,S.Graslund,M.Hammarstrom, A.Johansson,I.Johansson,T.Kotenyova,L.Lehtio,M.E.Nilsson,P.Nordlund, T.Nyman,K.Olesen,C.Persson,J.Sagemark,A.G.Thorsell,L.Tresaugues, S.Van Den Berg,J.Weigelt,M.Welin,M.Wikstrom,M.Wisniewska,H.Schuler, Structural Genomics Consortium (Sgc)
|
|
Key ref:
|
 |
T.Karlberg
et al.
Human tyrosine aminotransferase.
To be published,
.
|
 |
|
Date:
|
 |
|
27-Jul-08
|
Release date:
|
19-Aug-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P17735
(ATTY_HUMAN) -
Tyrosine aminotransferase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
454 a.a.
388 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.5
- tyrosine transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Phenylalanine and Tyrosine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-tyrosine + 2-oxoglutarate = 3-(4-hydroxyphenyl)pyruvate + L-glutamate
|
 |
 |
 |
 |
 |
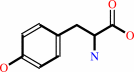 L-tyrosine
L-tyrosine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
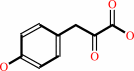 3-(4-hydroxyphenyl)pyruvate
3-(4-hydroxyphenyl)pyruvate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

