
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.1.14.11.30
- hypoxia-inducible factor-asparagine dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-asparaginyl-[hypoxia-inducible factor alpha subunit] + 2-oxoglutarate + O2 = (3S)-3-hydroxy-L-asparaginyl-[hypoxia-inducible factor alpha subunit] + succinate + CO2
|
 |
 |
 |
 |
 |
L-asparaginyl-[hypoxia-inducible factor alpha subunit]
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
(3S)-3-hydroxy-L-asparaginyl-[hypoxia-inducible factor alpha subunit]
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+); L-ascorbate
|
 |
 |
 |
 |
 |
Fe(2+)
|
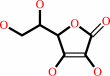 L-ascorbate
L-ascorbate
|
|
 |
 |
Enzyme class 3:
|
 |
Chain A:
E.C.1.14.11.n4
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:25971-25978
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH).
|
|
K.S.Hewitson,
S.L.Holmes,
D.Ehrismann,
A.P.Hardy,
R.Chowdhury,
C.J.Schofield,
M.A.McDonough.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A 2-His-1-carboxylate triad of iron binding residues is present in many non-heme
iron oxygenases including the Fe(II) and 2-oxoglutarate (2OG)-dependent
dioxygenases. Three variants (D201A, D201E, and D201G) of the iron binding
Asp-201 residue of an asparaginyl hydroxylase, factor inhibiting HIF (FIH), were
made and analyzed. FIH-D201A and FIH-D201E did not catalyze asparaginyl
hydroxylation, but in the presence of a reducing agent, they displayed enhanced
2OG turnover when compared with wild-type FIH. Turnover of 2OG by FIH-D201A was
significantly stimulated by the addition of HIF-1alpha(786-826) peptide. Like
FIH-D201A and D201E, the D201G variant enhanced 2OG turnover but rather
unexpectedly catalyzed asparaginyl hydroxylation. Crystal structures of the
FIH-D201A and D201G variants in complex with Fe(II)/Zn(II), 2OG, and
HIF-1alpha(786-826/788-806) implied that only two FIH-based residues (His-199
and His-279) are required for metal binding. The results indicate that variation
of 2OG-dependent dioxygenase iron-ligating residues as a means of functional
assignment should be treated with caution. The results are of mechanistic
interest in the light of recent biochemical and structural analyses of non-heme
iron and 2OG-dependent halogenases that are similar to the FIH-D201A/G variants
in that they use only two His-residues to ligate iron.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
The reaction catalyzed by wild-type FIH (and FIH-D201G).
hydroxylation of the β-carbon of Asn-803 within the C-terminal
transactivation domain of HIF-1α. The reaction requires Fe(II)
as a cofactor and dioxygen and 2OG as co-substrates that are
converted to succinate and carbon dioxide concomitant with
hydroxylation of HIF-1α substrate. For the FIH-D201A/E
variants, 2OG decarboxylation is uncoupled from the
hydroxylation of HIF-1α substrate and is stimulated by the
presence of a reducing agent (ascorbate or DTT).
|
 |
Figure 2.
Insights from crystal structures of FIH-D201A and FIH-D201G.
a, stereo view of the iron binding site of the
FIH-D201A·Fe(II)·2OG·HIF-1α[786–826]
complex. Experimental electron density OMIT map (F[o] - F[c])
contoured to 5σ represented as blue mesh (electron density is
carved out around the residues and ligands displayed for
clarity). The unanticipated electron density adjacent to the
iron was provisionally modeled as (bi)carbonate (see
“Results” for discussion). b, comparison of the wild-type
FIH·Fe(II)·2OG·HIF-1α[786–826] complex
(PDB ID 1H2L) with the
FIH-D201A·Fe(II)·2OG·HIF-1α[786–826]
complex (wild-type FIH (blue) in complex with HIF substrate
(cyan) and FIH-D201A (yellow) in complex with HIF substrate
(magenta)). This figure emphasizes several important
interactions between wild-type FIH and HIF-1α that are lost in
the FIH-D201A complex: wild-type FIH Asp-201 and HIF-1α Asn-803
main chain nitrogen (yellow dash), wild-type FIH Gln-239 and
HIF-1α Asn-803 side chain, and wild-type FIH Trp-296 and
HIF-1α Val-802. Note the presence of the assigned sulfate ion
(orange and red) in the FIH-D201A structure apparently replacing
the carboxylate of the HIF-1α Glu-801 in the wild-type
FIH·Fe(II)·2OG·HIF-1α[786–826] complex,
which is also observed in uncomplexed wild-type FIH structure
(PDB 1H2N, not shown). c, stereo views from the crystal
structures of 2OG-dependent halogenase, SyrB2 (pink)
superimposed on the FIH-D201A variant (yellow). The FIH-D201A
variant shares the same HXA... H motif as SyrB2 (PDB ID 2FCT);
however, the FIH-D201A apparently does not provide enough space
for a chloride ion to complete octahedral coordination to the
Fe(II), which could explain why FIH-D201A does not have
halogenase activity toward HIF-1α under our assay conditions.
Distances between the FIH-D201A Ala-201 Cβ methyl group and
Fe(II) in each of the structures are shown as black dashed lines
to emphasize this point. d, stereo view ball-and-stick
representation of the
FIH-D201G·Zn(II)·2OG·HIF-1α[786–826]
complex metal binding site. Experimental electron density 2F[o]
- F[c] contoured to 1.0σ represented as blue mesh (electron
density is carved out around the residues and ligands displayed
for clarity). FIH and 2OG are colored green, HIF-1α[786–826]
is colored yellow, Zn(II) is colored as a gray sphere, and the
Zn(II) bound water is colored as a red sphere. Wat, water.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2008,
283,
25971-25978)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Yan,
M.Kong,
and
Y.H.Chen
(2011).
Prevention of apoptosis by the interaction between FIH1 and Bax.
|
| |
Mol Cell Biochem,
348,
1-9.
|
 |
|
|
|
|
 |
F.S.Lee,
and
M.J.Percy
(2011).
The HIF pathway and erythrocytosis.
|
| |
Annu Rev Pathol,
6,
165-192.
|
 |
|
|
|
|
 |
M.Yang,
W.Ge,
R.Chowdhury,
T.D.Claridge,
H.B.Kramer,
B.Schmierer,
M.A.McDonough,
L.Gong,
B.M.Kessler,
P.J.Ratcliffe,
M.L.Coleman,
and
C.J.Schofield
(2011).
Asparagine and Aspartate Hydroxylation of the Cytoskeletal Ankyrin Family Is Catalyzed by Factor-inhibiting Hypoxia-inducible Factor.
|
| |
J Biol Chem,
286,
7648-7660.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.R.Rose,
E.C.Woon,
G.L.Kingham,
O.N.King,
J.Mecinović,
I.J.Clifton,
S.S.Ng,
J.Talib-Hardy,
U.Oppermann,
M.A.McDonough,
and
C.J.Schofield
(2010).
Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches.
|
| |
J Med Chem,
53,
1810-1818.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.N.King,
X.S.Li,
M.Sakurai,
A.Kawamura,
N.R.Rose,
S.S.Ng,
A.M.Quinn,
G.Rai,
B.T.Mott,
P.Beswick,
R.J.Klose,
U.Oppermann,
A.Jadhav,
T.D.Heightman,
D.J.Maloney,
C.J.Schofield,
and
A.Simeonov
(2010).
Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors.
|
| |
PLoS One,
5,
e15535.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Hahn,
I.Wegener,
A.Burrells,
J.Böse,
A.Wolf,
C.Erck,
D.Butler,
C.J.Schofield,
A.Böttger,
and
A.Lengeling
(2010).
Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation.
|
| |
PLoS One,
5,
e13769.
|
 |
|
|
|
|
 |
H.J.Kulik,
L.C.Blasiak,
N.Marzari,
and
C.L.Drennan
(2009).
First-principles study of non-heme Fe(II) halogenase SyrB2 reactivity.
|
| |
J Am Chem Soc,
131,
14426-14433.
|
 |
|
|
|
|
 |
L.J.Cliffe,
R.Kieft,
T.Southern,
S.R.Birkeland,
M.Marshall,
K.Sweeney,
and
R.Sabatini
(2009).
JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes.
|
| |
Nucleic Acids Res,
37,
1452-1462.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

