 |
PDBsum entry 3cl6

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.2.5
- allantoinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-allantoin + H2O = allantoate + H+
|
 |
 |
 |
 |
 |
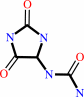 (S)-allantoin
(S)-allantoin
|
+
|
H2O
|
=
|
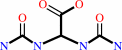 allantoate
allantoate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:23295-23304
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Logical Identification of an Allantoinase Analog (puuE) Recruited from Polysaccharide Deacetylases.
|
|
I.Ramazzina,
L.Cendron,
C.Folli,
R.Berni,
D.Monteverdi,
G.Zanotti,
R.Percudani.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The hydrolytic cleavage of the hydantoin ring of allantoin, catalyzed by
allantoinase, is required for the utilization of the nitrogen present in
purine-derived compounds. The allantoinase gene (DAL1), however, is missing in
many completely sequenced organisms able to use allantoin as a nitrogen source.
Here we show that an alternative allantoinase gene (puuE) can be precisely
identified by analyzing its logic relationship with three other genes of the
pathway. The novel allantoinase is annotated in structure and sequence data
bases as polysaccharide deacetylase for its homology with enzymes that catalyze
hydrolytic reactions on chitin or peptidoglycan substrates. The recombinant PuuE
protein from Pseudomonas fluorescens exhibits metal-independent allantoinase
activity and stereospecificity for the S enantiomer of allantoin. The crystal
structures of the protein and of protein-inhibitor complexes reveal an overall
similarity with the polysaccharide deacetylase beta/alpha barrel and remarkable
differences in oligomeric assembly and active site geometry. The conserved
Asp-His-His metal-binding triad is replaced by Glu-His-Trp, a configuration that
is distinctive of PuuE proteins within the protein family. An extra domain at
the top of the barrel offers a scaffold for protein tetramerization and forms a
small substrate-binding cleft by hiding the large binding groove of
polysaccharide deacetylases. Substrate positioning at the active site suggests
an acid/base mechanism of catalysis in which only one member of the catalytic
pair of polysaccharide deacetylases has been conserved. These data provide a
structural rationale for the shifting of substrate specificity that occurred
during evolution.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIGURE 3. Sequence and structure of PuuE. A, the PuuE
sequence is aligned with a hypothetical polysaccharide
deacetylase from P. aeruginosa (Protein Data Bank code 1Z7A), a
putative chitin deacetylase from S. pombe (SpCDA), a
peptidoglycan deacetylase from B. subtilis (Protein Data Bank
code 1W17), acetyl xylan esterases from S. lividans (Protein
Data Bank code 2CC0), and a chitin deacetylase from C.
lindemuthianum (Protein Data Bank code 2IW0). Secondary
structure elements deriving from Protein Data Bank coordinates
are drawn over the alignment. Residues involved in metal-binding
or in catalysis (38) are denoted by cyan (metal-binding) and
magenta (catalysis) arrowheads. B, cartoon drawing of the
structure of the PuuE monomer alongside its topological
representation. The helices are colored red, and the strands are
blue, except for helix  1(orange) and strands
β6 and β7(light blue), which do not fit the canonical PsDA
fold. C, cartoon drawing of the PuuE tetramer. Subunits A and C
are colored as in B; subunits B and D are colored green. 1(orange) and strands
β6 and β7(light blue), which do not fit the canonical PsDA
fold. C, cartoon drawing of the PuuE tetramer. Subunits A and C
are colored as in B; subunits B and D are colored green.
|
 |
Figure 4.
FIGURE 4. Substrate binding and reaction mechanism of PuuE.
A, surface drawing of hydantoin-bound PuuE (left panel) compared
with a zinc-bound chitin deacetylase (Protein Data Bank code
2IW0; right panel). The inserted segments IS1 and IS2 of PuuE
are colored orange and light blue, respectively. The positions
of sugar-binding subsites in 2IW0 are indicated. B, stereo view
comparison of the metal-binding site of 2IW0 (cyan) and the
corresponding region of PuuE (yellow). Metal-binding residues of
2IW0 and superimposable residues of PuuE are shown as sticks. C,
the PuuE active site in complex with hydantoin. Key residues and
ligands are represented in stick mode. D, proposed catalytic
mechanism for (S)-allantoin hydrolysis.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2008,
283,
23295-23304)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Ramazzina,
R.Costa,
L.Cendron,
R.Berni,
A.Peracchi,
G.Zanotti,
and
R.Percudani
(2010).
An aminotransferase branch point connects purine catabolism to amino acid recycling.
|
| |
Nat Chem Biol,
6,
801-806.
|
 |
|
|
|
|
 |
S.D.Pope,
L.L.Chen,
and
V.Stewart
(2009).
Purine utilization by Klebsiella oxytoca M5al: genes for ring-oxidizing and -opening enzymes.
|
| |
J Bacteriol,
191,
1006-1017.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

