 |
PDBsum entry 3cb0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3cb0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Cobr
|
|
Structure:
|
 |
4-hydroxyphenylacetate 3-monooxygenase. Chain: a, b, c, d. Synonym: cobr, corrin reductase. Engineered: yes
|
|
Source:
|
 |
Brucella melitensis. Organism_taxid: 29459. Gene: cobr. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.60Å
|
R-factor:
|
0.197
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
A.D.Lawrence,M.J.Warren,R.W.Pickersgill
|
Key ref:

|
 |
A.D.Lawrence
et al.
(2008).
Identification, characterization, and structure/function analysis of a corrin reductase involved in adenosylcobalamin biosynthesis.
J Biol Chem,
283,
10813-10821.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
21-Feb-08
|
Release date:
|
11-Mar-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8YHT7
(Q8YHT7_BRUME) -
4-hydroxyphenylacetate 3-monooxygenase from Brucella melitensis biotype 1 (strain 16M / ATCC 23456 / NCTC 10094)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
173 a.a.
166 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.14.9
- 4-hydroxyphenylacetate 3-monooxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxyphenylacetate + FADH2 + O2 = 3,4-dihydroxyphenylacetate + FAD + H2O + H+
|
 |
 |
 |
 |
 |
 4-hydroxyphenylacetate
4-hydroxyphenylacetate
|
+
|
FADH2
Bound ligand (Het Group name = )
matches with 58.49% similarity
|
+
|
 O2
O2
|
=
|
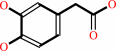 3,4-dihydroxyphenylacetate
3,4-dihydroxyphenylacetate
|
+
|
 FAD
FAD
|
+
|
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
 FAD
FAD
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:10813-10821
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Identification, characterization, and structure/function analysis of a corrin reductase involved in adenosylcobalamin biosynthesis.
|
|
A.D.Lawrence,
E.Deery,
K.J.McLean,
A.W.Munro,
R.W.Pickersgill,
S.E.Rigby,
M.J.Warren.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Vitamin B(12), the antipernicious anemia factor, is the cyano derivative of
adenosylcobalamin, which is one of nature's most complex coenzymes.
Adenosylcobalamin is made along one of two similar yet distinct metabolic
pathways, which are referred to as the aerobic and anaerobic routes. The aerobic
pathway for cobalamin biosynthesis proceeds via cobalt insertion into a
ring-contracted macrocycle, which is closely followed by adenosylation of the
cobalt ion. An important prerequisite for adenosylation is the reduction of the
centrally chelated metal from Co(II) to a highly nucleophilic Co(I) form. We
have cloned a gene, cobR, encoding a biosynthetic enzyme with this co(II)rrin
reductase activity from Brucella melitensis. The protein has been overproduced,
and the resulting flavoprotein has been purified, characterized, and
crystallized and its structure determined to 1.6A resolution. Kinetic and EPR
analysis reveals that the enzyme proceeds via a semiquinone form. It is proposed
that CobR may interact with the adenosyltransferase to overcome the large
thermodynamic barrier required for co(II)rrin reduction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. Biosynthesis of vitamin B[12], highlighting the
transformation of cob(II)yrinic acid a,c-diamide into
adenonsylcob(III)yrinic acid a,c-diamide. After the insertion of
cobalt into the contracted ring system by CobNST, the central
cobalt ion of cobyrinic acid a,c-diamide is reduced to the Co(I)
form by CobR, which acts as a nucleophile for the adenosylation
process.
|
 |
Figure 6.
FIGURE 6. Crystal structure of CobR. a, schematic
representation of the homodimer architecture of CobR with FMN
shown in stick representation. The si face of the FMN is
presented to the large central cavity. b,  [A] weighted 2F[obs] -
F[calc] Fourier synthesis showing the quality of the electron
density corresponding to the isoalloxazine ring of the bound
FMN; the electron density is contoured at 1 [A] weighted 2F[obs] -
F[calc] Fourier synthesis showing the quality of the electron
density corresponding to the isoalloxazine ring of the bound
FMN; the electron density is contoured at 1  (blue mesh). c,
schematic representation showing the superimposition of flavin
reductase (Phe(A2)) and CobR, which brings the isoalloxazine
rings of the flavins shown in stick representation into close
structural alignment. The position of (blue mesh). c,
schematic representation showing the superimposition of flavin
reductase (Phe(A2)) and CobR, which brings the isoalloxazine
rings of the flavins shown in stick representation into close
structural alignment. The position of  -helix -helix  4 of
CobR precludes the binding of adenosine at the corresponding
position in CobR, and there is no adjacent binding pocket that
can accommodate the adenosine. The NAD bound by flavin reductase
can be readily accommodated in CobR. d, more detailed view of
FMN and proposed NAD binding in CobR showing some key
interactions with FMN and NAD. The measured distance between the
modeled NAD (nicotinamide) C-5 and the experimentally observed
isoalloxazine ring N-5 of 3.2 Å is consistent with hydride
transfer from C-5 of NADH to N-5 of FMN. Produced using PyMol
(17). 4 of
CobR precludes the binding of adenosine at the corresponding
position in CobR, and there is no adjacent binding pocket that
can accommodate the adenosine. The NAD bound by flavin reductase
can be readily accommodated in CobR. d, more detailed view of
FMN and proposed NAD binding in CobR showing some key
interactions with FMN and NAD. The measured distance between the
modeled NAD (nicotinamide) C-5 and the experimentally observed
isoalloxazine ring N-5 of 3.2 Å is consistent with hydride
transfer from C-5 of NADH to N-5 of FMN. Produced using PyMol
(17).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2008,
283,
10813-10821)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.N.Webb,
J.W.Ballinger,
E.Kim,
S.M.Belchik,
K.S.Lam,
B.Youn,
M.S.Nissen,
L.Xun,
and
C.Kang
(2010).
Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100.
|
| |
J Biol Chem,
285,
2014-2027.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.B.Parsons,
A.D.Lawrence,
K.J.McLean,
A.W.Munro,
S.E.Rigby,
and
M.J.Warren
(2010).
Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment.
|
| |
PLoS One,
5,
e14009.
|
 |
|
|
|
|
 |
P.E.Mera,
and
J.C.Escalante-Semerena
(2010).
Multiple roles of ATP:cob(I)alamin adenosyltransferases in the conversion of B12 to coenzyme B12.
|
| |
Appl Microbiol Biotechnol,
88,
41-48.
|
 |
|
|
|
|
 |
P.E.Mera,
and
J.C.Escalante-Semerena
(2010).
Dihydroflavin-driven adenosylation of 4-coordinate Co(II) corrinoids: are cobalamin reductases enzymes or electron transfer proteins?
|
| |
J Biol Chem,
285,
2911-2917.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

