 |
PDBsum entry 3c20

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.2.4
- aspartate kinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Lysine biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate + ATP = 4-phospho-L-aspartate + ADP
|
 |
 |
 |
 |
 |
 L-aspartate
L-aspartate
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
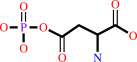 4-phospho-L-aspartate
4-phospho-L-aspartate
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:16216-16225
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis for allosteric inhibition of a threonine-sensitive aspartokinase.
|
|
X.Liu,
A.G.Pavlovsky,
R.E.Viola.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The commitment step to the aspartate pathway of amino acid biosynthesis is the
phosphorylation of aspartic acid catalyzed by aspartokinase (AK). Most
microorganisms and plants have multiple forms of this enzyme, and many of these
isofunctional enzymes are subject to feedback regulation by the end products of
the pathway. However, the archeal species Methanococcus jannaschii has only a
single, monofunctional form of AK. The substrate l-aspartate binds to this
recombinant enzyme in two different orientations, providing the first structural
evidence supporting the relaxed regiospecificity previously observed with
several alternative substrates of Escherichia coli AK ( Angeles, T. S., Hunsley,
J. R., and Viola, R. E. (1992) Biochemistry 31, 799-805 ). Binding of the
nucleotide substrate triggers significant domain movements that result in a more
compact quaternary structure. In contrast, the highly cooperative binding of the
allosteric regulator l-threonine to multiple sites on this dimer of dimers leads
to an open enzyme structure. A comparison of these structures supports a
mechanism for allosteric regulation in which the domain movements induced by
threonine binding causes displacement of the substrates from the enzyme,
resulting in a relaxed, inactive conformation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIGURE 3. Nucleotide-induced domain closure in mjAK. A, an
overlay of the ternary complex (yellow ribbons) on the binary
complex (blue ribbons) shows that binding of the ATP analog
AMP-PNP induces a 12.5° rotation around the hinge bending
region (green) of the regulatory domain (light blue) toward the
kinase domain (dark blue). B, an expansion of the active site
showing the movement of latch loop I into position to form
binding interactions with the ribose ring of AMP-PNP and the
closing of latch loop II to complete the hydrophobic pocket of
adenine ring binding.
|
 |
Figure 4.
FIGURE 4. The mjAK/L-threonine structure reveals two sets
of threonine binding sites. Each L-threonine is positioned in
the binding site by interactions between its functional groups
and the enzyme. Inset A, the binding modes of two threonines at
the A-B dimer interface site. This inset is rotated by 90°
to provide a clearer view of the two bound threonines. Inset B,
the binding of a single threonine with lower occupancy at the
C-D dimer interface site. This inset is rotated by 180° to
show the binding interactions at this site. Inset C,
representative binding of threonine at the weaker secondary
sites in each monomer, located adjacent to the active site.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2008,
283,
16216-16225)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Chaitanya,
B.Babajan,
C.M.Anuradha,
M.Naveen,
C.Rajasekhar,
P.Madhusudana,
and
C.S.Kumar
(2010).
Exploring the molecular basis for selective binding of Mycobacterium tuberculosis Asp kinase toward its natural substrates and feedback inhibitors: a docking and molecular dynamics study.
|
| |
J Mol Model,
16,
1357-1367.
|
 |
|
|
|
|
 |
M.F.Mabanglo,
H.L.Schubert,
M.Chen,
C.P.Hill,
and
C.D.Poulter
(2010).
X-ray structures of isopentenyl phosphate kinase.
|
| |
ACS Chem Biol,
5,
517-527.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Dellas,
and
J.P.Noel
(2010).
Mutation of archaeal isopentenyl phosphate kinase highlights mechanism and guides phosphorylation of additional isoprenoid monophosphates.
|
| |
ACS Chem Biol,
5,
589-601.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Yoshida,
T.Tomita,
H.Kono,
S.Fushinobu,
T.Kuzuyama,
and
M.Nishiyama
(2009).
Crystal structures of the regulatory subunit of Thr-sensitive aspartate kinase from Thermus thermophilus.
|
| |
FEBS J,
276,
3124-3136.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.C.Lo,
C.A.Bonner,
G.Xie,
M.D'Souza,
and
R.A.Jensen
(2009).
Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways.
|
| |
Microbiol Mol Biol Rev,
73,
594-651.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

