 |
PDBsum entry 3a3d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.4.16.4
- serine-type D-Ala-D-Ala carboxypeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-alanyl-D-alanine + H2O = 2 D-alanine
|
 |
 |
 |
 |
 |
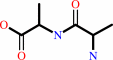
|
+
|
|
=
|
2
×
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.4.21.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
396:634-645
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae.
|
|
F.Kawai,
T.B.Clarke,
D.I.Roper,
G.J.Han,
K.Y.Hwang,
S.Unzai,
E.Obayashi,
S.Y.Park,
J.R.Tame.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We have determined high-resolution apo crystal structures of two low molecular
weight penicillin-binding proteins (PBPs), PBP4 and PBP5, from Haemophilus
influenzae, one of the most frequently found pathogens in the upper respiratory
tract of children. Novel beta-lactams with notable antimicrobial activity have
been designed, and crystal structures of PBP4 complexed with ampicillin and two
of the novel molecules have also been determined. Comparing the apo form with
those of the complexes, we find that the drugs disturb the PBP4 structure and
weaken X-ray diffraction, to very different extents. PBP4 has recently been
shown to act as a sensor of the presence of penicillins in Pseudomonas
aeruginosa, and our models offer a clue to the structural basis for this effect.
Covalently attached penicillins press against a phenylalanine residue near the
active site and disturb the deacylation step. The ready inhibition of PBP4 by
beta-lactams compared to PBP5 also appears to be related to the weaker
interactions holding key residues in a catalytically competent position.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Zamorano,
T.M.Reeve,
L.Deng,
C.Juan,
B.Moyá,
G.Cabot,
D.J.Vocadlo,
B.L.Mark,
and
A.Oliver
(2010).
NagZ inactivation prevents and reverts beta-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa.
|
| |
Antimicrob Agents Chemother,
54,
3557-3563.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

