 |
PDBsum entry 2zy2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.1
- aspartate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate
|
 |
 |
 |
 |
 |
 L-aspartate
L-aspartate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 oxaloacetate
oxaloacetate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.4.1.1.12
- aspartate 4-decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate + H+ = L-alanine + CO2
|
 |
 |
 |
 |
 |
 L-aspartate
L-aspartate
|
+
|
H(+)
|
=
|
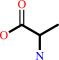 L-alanine
L-alanine
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
17:517-529
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure, assembly, and mechanism of a PLP-dependent dodecameric L-aspartate beta-decarboxylase.
|
|
H.J.Chen,
T.P.Ko,
C.Y.Lee,
N.C.Wang,
A.H.Wang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The type-I PLP enzyme l-aspartate beta-decarboxylase converts aspartate to
alanine and CO(2). Similar to the homodimeric aminotransferases, its protein
subunit comprises a large and a small domain, of 410 and 120 residues,
respectively. The crystal structure reveals a dodecamer made of six identical
dimers arranged in a truncated tetrahedron whose assembly involves tetramer and
hexamer as intermediates. The additional helical motifs I and II participate in
the oligomer formation. Triple mutations of S67R/Y68R/M69R or S67E/Y68E/M69E in
motif I produced an inactive dimer. The PLP is bound covalently to Lys315 in the
active site, while its phosphate group interacts with a neighboring Tyr134.
Removal of the bulky side chain of Arg37, which overhangs the PLP group,
improved the substrate affinity. Mutations in flexible regions produced the more
active K17A and the completely inactive R487A. The structure also suggests that
substrate binding triggers conformational changes essential for catalyzing the
reaction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. The pH Dependence of the Dodecamer Assembly of AsdA
(A) The distribution of molecular size under various pH
conditions was assessed using AUC. Parameters used in
calculating the sedimentation coefficients are viscosity =
0.01048 and density = 1.01029.
(B) The curve at pH 7.5 was
converted to represent mass distribution. The corresponding
masses of dimer and dodecamer were calculated using frictional
coefficients f = 1.32–1.35, and those of the intermediates
(inset; probably tetramer and hexamer) using f = 1.21–1.25.
(C) Areas under the curves in (A) were integrated to show
different proportions of oligomers in various pH conditions. The
ratio of dimer: intermediates: dodecamer was roughly 1:1:2 at pH
8.
(D) Under higher ionic strength of KCl, more dodecamers
were dissociated into dimers at pH 7.
(E) The proposed
assembly mechanism of ASD starts from monomer to dimer, followed
by tetramer and two kinds of hexamer. Finally the two hexamers
dock together, face to face, to form the spherical dodecamer.
|
 |
Figure 5.
Figure 5. The Active Site Structure
(A) Stereoscopic
view of the PLP binding site shows the surroundings of the bound
cofactor (yellow). The phosphate group and the protonated
nitrogen are salt-bridged to Arg323 and Asp286. The cyan Tyr134
is from another monomer.
(B) The active-site regions of two
different subunits are superimposed. The L and S domains are
colored cyan and green. N-terminal helix α1, and loops α6-α7
and βY-βZ, which contain Lys17, Tyr134, and Arg487,
respectively, and the covering helix α9 are highlighted in
purple, pink, red, and blue. Primed residue numbers are for the
more open molecule.
(C) Surface presentation of the closed
active site. Positive and negative charge potentials are
indicated by blue and red colors. The loops and helices forming
the active-site entrance are also shown.
(D) Surface of the
more open active site. Intrinsic flexibility of domains,
secondary structure elements, and loops should have caused the
structural difference.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(2009,
17,
517-529)
copyright 2009.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

