 |
PDBsum entry 2wd4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2wd4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.11
- L-ascorbate peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ascorbate + H2O2 = L-dehydroascorbate + 2 H2O
|
 |
 |
 |
 |
 |
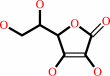 L-ascorbate
L-ascorbate
|
+
|
 H2O2
H2O2
|
=
|
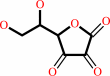 L-dehydroascorbate
L-dehydroascorbate
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
TBV)
matches with 83.67% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
48:4738-4746
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Evidence for heme oxygenase activity in a heme peroxidase.
|
|
S.K.Badyal,
G.Eaton,
S.Mistry,
Z.Pipirou,
J.Basran,
C.L.Metcalfe,
A.Gumiero,
S.Handa,
P.C.Moody,
E.L.Raven.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The heme peroxidase and heme oxygenase enzymes share a common heme prosthetic
group but catalyze fundamentally different reactions, the first being
H(2)O(2)-dependent oxidation of substrate using an oxidized Compound I
intermediate, and the second O(2)-dependent degradation of heme. It has been
proposed that these enzymes utilize a common reaction intermediate, a ferric
hydroperoxide species, that sits at a crossroads in the mechanism and beyond
which there are two mutually exclusive mechanistic pathways. Here, we present
evidence to support this proposal in a heme peroxidase. Hence, we describe
kinetic data for a variant of ascorbate peroxidase (W41A) which reacts slowly
with tert-butyl hydroperoxide and does not form the usual peroxidase Compound I
intermediate; instead, structural data show that a product is formed in which
the heme has been cleaved at the alpha-meso position, analogous to the heme
oxygenase mechanism. We interpret this to mean that the Compound I (peroxidase)
pathway is shut down, so that instead the reaction intermediate diverts through
the alternative (heme oxygenase) route. A mechanism for formation of the product
is proposed and discussed in the light of what is known about the heme oxygenase
reaction mechanism.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Ishikawa,
N.Tajima,
H.Nishikawa,
Y.Gao,
M.Rapolu,
H.Shibata,
Y.Sawa,
and
S.Shigeoka
(2010).
Euglena gracilis ascorbate peroxidase forms an intramolecular dimeric structure: its unique molecular characterization.
|
| |
Biochem J,
426,
125-134.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

