 |
PDBsum entry 2vbv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.161
- CTP-dependent riboflavin kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
riboflavin + CTP = CDP + FMN + H+
|
 |
 |
 |
 |
 |
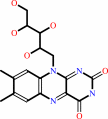 riboflavin
riboflavin
|
+
|
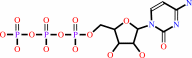 CTP
CTP
|
=
|
CDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
FMN
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
15:1577-1590
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
A CTP-dependent archaeal riboflavin kinase forms a bridge in the evolution of cradle-loop barrels.
|
|
M.Ammelburg,
M.D.Hartmann,
S.Djuranovic,
V.Alva,
K.K.Koretke,
J.Martin,
G.Sauer,
V.Truffault,
K.Zeth,
A.N.Lupas,
M.Coles.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Proteins of the cradle-loop barrel metafold are formed by duplication of a
conserved betaalphabeta-element, suggesting a common evolutionary origin from an
ancestral group of nucleic acid-binding proteins. The basal fold within this
metafold, the RIFT barrel, is also found in a wide range of enzymes, whose
homologous relationship with the nucleic acid-binding group is unclear. We have
characterized a protein family that is intermediate in sequence and structure
between the basal group of cradle-loop barrels and one family of RIFT-barrel
enzymes, the riboflavin kinases. We report the structure, substrate-binding
mode, and catalytic activity for one of these proteins, Methanocaldococcus
jannaschii Mj0056, which is an archaeal riboflavin kinase. Mj0056 is unusual in
utilizing CTP rather than ATP as the donor nucleotide, and sequence conservation
in the relevant residues suggests that this is a general feature of archaeal
riboflavin kinases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Cluster Map of Mj0065 Homologs
The map,
obtained with CLANS (see Experimental Procedures), shows Mj0056
in the context of double-psi barrels (AAA N domains, double-psi
barrel enzymes), swapped-hairpin barrels (AbrB superfamily), and
RIFT barrels (PhS018 group, riboflavin kinases, riboflavin
synthases).
|
 |
Figure 8.
Figure 8. Structural Comparison of Archaeal and
Bacterial/Eukaryotic RFKs
Mj0056-MgCDP-FMN (A) is compared
to HsRFK-MgADP-FMN (1Q9S) (B). A superposition of the nucleotide
binding sites (C) illustrates how the absence of α1′ in HsRFK
accommodates the larger adenosine moiety. The superimposition in
(D) shows the FMN binding site, highlighting the structural
equivalence of α1 in Mj0056 to the C-terminal extension in
HsRFK.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(2007,
15,
1577-1590)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Shi,
B.Chitturi,
and
N.V.Grishin
(2009).
ProSMoS server: a pattern-based search using interaction matrix representation of protein structures.
|
| |
Nucleic Acids Res,
37,
W526-W531.
|
 |
|
|
|
|
 |
V.Alva,
S.Dunin-Horkawicz,
M.Habeck,
M.Coles,
and
A.N.Lupas
(2009).
The GD box: a widespread noncontiguous supersecondary structural element.
|
| |
Protein Sci,
18,
1961-1966.
|
 |
|
|
|
|
 |
V.Y.Yatsyshyn,
O.P.Ishchuk,
A.Y.Voronovsky,
D.V.Fedorovych,
and
A.A.Sibirny
(2009).
Production of flavin mononucleotide by metabolically engineered yeast Candida famata.
|
| |
Metab Eng,
11,
163-167.
|
 |
|
|
|
|
 |
V.Alva,
K.K.Koretke,
M.Coles,
and
A.N.Lupas
(2008).
Cradle-loop barrels and the concept of metafolds in protein classification by natural descent.
|
| |
Curr Opin Struct Biol,
18,
358-365.
|
 |
|
|
|
|
 |
Z.Mashhadi,
H.Zhang,
H.Xu,
and
R.H.White
(2008).
Identification and characterization of an archaeon-specific riboflavin kinase.
|
| |
J Bacteriol,
190,
2615-2618.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

