 |
PDBsum entry 2qd2

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
2qd2
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.98.1.1
- protoporphyrin ferrochelatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
heme b + 2 H+ = protoporphyrin IX + Fe2+
|
 |
 |
 |
 |
 |
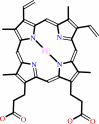 heme b
heme b
|
+
|
2
×
H(+)
|
=
|
 protoporphyrin IX
protoporphyrin IX
|
+
|
Fe(2+)
Bound ligand (Het Group name = )
matches with 97.67% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
373:1006-1016
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase.
|
|
A.E.Medlock,
T.A.Dailey,
T.A.Ross,
H.A.Dailey,
W.N.Lanzilotta.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ferrochelatase (protoheme ferrolyase, EC 4.99.1.1) is the terminal enzyme in
heme biosynthesis and catalyzes the insertion of ferrous iron into
protoporphyrin IX to form protoheme IX (heme). Due to the many critical roles of
heme, synthesis of heme is required by the vast majority of organisms. Despite
significant investigation of both the microbial and eukaryotic enzyme, details
of metal chelation remain unidentified. Here we present the first structure of
the wild-type human enzyme, a lead-inhibited intermediate of the wild-type
enzyme with bound metallated porphyrin macrocycle, the product bound form of the
enzyme, and a higher resolution model for the substrate-bound form of the E343K
variant. These data paint a picture of an enzyme that undergoes significant
changes in secondary structure during the catalytic cycle. The role that these
structural alterations play in overall catalysis and potential protein-protein
interactions with other proteins, as well as the possible molecular basis for
these changes, is discussed. The atomic details and structural rearrangements
presented herein significantly advance our understanding of the substrate
binding mode of ferrochelatase and reveal new conformational changes in a
structurally conserved pi-helix that is predicted to have a central role in
product release.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Occupancy of the active site for WT1 and R115L. (a)
Overlay of cholate residues and the side-chains of H263, R115
and M76 of the WT1 model with the model previously reported for
the R115L variant of human ferrochelatase. (b) Heme molecule
modeled into the density present in the active site of monomer B
of the WT1 data with side-chains of M76 below and H263 above the
macrocycle ring. All atoms in the WT1 model are represented as
sticks with nitrogen, oxygen, carbon, sulfur, and iron atoms
colored blue, red, tan, cyan, and black, respectively. All atoms
in the model previously reported for R115L variant are colored
green in (a). In all cases the 2F[o]–F[c] composite omit map
was generated using the simulated annealing protocol and is
contoured at 1 σ (purple and green cage in (a) and (b),
respectively).
|
 |
Figure 5.
Figure 5. Electrostatic surface potential showing the active
site region for the (a) wild-type, (b) the substrate bound, and
(c) heme bound human ferrochelatase. For clarity, the upper lip
and π-helix regions are highlighted. The Figure was generated
with PyMOL [http://pymol.sourceforge.net/].
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Elsevier:
J Mol Biol
(2007,
373,
1006-1016)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.V.Romão,
D.Ladakis,
S.A.Lobo,
M.A.Carrondo,
A.A.Brindley,
E.Deery,
P.M.Matias,
R.W.Pickersgill,
L.M.Saraiva,
and
M.J.Warren
(2011).
Evolution in a family of chelatases facilitated by the introduction of active site asymmetry and protein oligomerization.
|
| |
Proc Natl Acad Sci U S A,
108,
97.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.D.Hansson,
T.Karlberg,
C.A.Söderberg,
S.Rajan,
M.J.Warren,
S.Al-Karadaghi,
S.E.Rigby,
and
M.Hansson
(2011).
Bacterial ferrochelatase turns human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase.
|
| |
J Biol Inorg Chem,
16,
235-242.
|
 |
|
|
|
|
 |
N.R.McIntyre,
R.Franco,
J.A.Shelnutt,
and
G.C.Ferreira
(2011).
Nickel(II) chelatase variants directly evolved from murine ferrochelatase: porphyrin distortion and kinetic mechanism.
|
| |
Biochemistry,
50,
1535-1544.
|
 |
|
|
|
|
 |
G.Layer,
J.Reichelt,
D.Jahn,
and
D.W.Heinz
(2010).
Structure and function of enzymes in heme biosynthesis.
|
| |
Protein Sci,
19,
1137-1161.
|
 |
|
|
|
|
 |
W.Chen,
H.A.Dailey,
and
B.H.Paw
(2010).
Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis.
|
| |
Blood,
116,
628-630.
|
 |
|
|
|
|
 |
A.E.Medlock,
M.Carter,
T.A.Dailey,
H.A.Dailey,
and
W.N.Lanzilotta
(2009).
Product release rather than chelation determines metal specificity for ferrochelatase.
|
| |
J Mol Biol,
393,
308-319.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Szefczyk,
M.N.Cordeiro,
R.Franco,
and
J.A.Gomes
(2009).
Molecular dynamics simulations of mouse ferrochelatase variants: what distorts and orientates the porphyrin?
|
| |
J Biol Inorg Chem,
14,
1119-1128.
|
 |
|
|
|
|
 |
R.E.Davidson,
C.J.Chesters,
and
J.D.Reid
(2009).
Metal ion selectivity and substrate inhibition in the metal ion chelation catalyzed by human ferrochelatase.
|
| |
J Biol Chem,
284,
33795-33799.
|
 |
|
|
|
|
 |
G.A.Hunter,
M.P.Sampson,
and
G.C.Ferreira
(2008).
Metal ion substrate inhibition of ferrochelatase.
|
| |
J Biol Chem,
283,
23685-23691.
|
 |
|
|
|
|
 |
T.Karlberg,
M.D.Hansson,
R.K.Yengo,
R.Johansson,
H.O.Thorvaldsen,
G.C.Ferreira,
M.Hansson,
and
S.Al-Karadaghi
(2008).
Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues.
|
| |
J Mol Biol,
378,
1074-1083.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

