 |
PDBsum entry 2pme

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.7.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.6.1.1.14
- glycine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Gly) + glycine + ATP = glycyl-tRNA(Gly) + AMP + diphosphate
|
 |
 |
 |
 |
 |
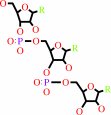 tRNA(Gly)
tRNA(Gly)
|
+
|
 glycine
glycine
|
+
|
 ATP
ATP
|
=
|
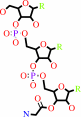 glycyl-tRNA(Gly)
glycyl-tRNA(Gly)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
104:9976-9981
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase.
|
|
W.Xie,
L.A.Nangle,
W.Zhang,
P.Schimmel,
X.L.Yang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Functional expansion of specific tRNA synthetases in higher organisms is well
documented. These additional functions may explain why dominant mutations in
glycyl-tRNA synthetase (GlyRS) and tyrosyl-tRNA synthetase cause
Charcot-Marie-Tooth (CMT) disease, the most common heritable disease of the
peripheral nervous system. At least 10 disease-causing mutant alleles of GlyRS
have been annotated. These mutations scatter broadly across the primary sequence
and have no apparent unifying connection. Here we report the structure of wild
type and a CMT-causing mutant (G526R) of homodimeric human GlyRS. The mutation
is at the site for synthesis of glycyl-adenylate, but the rest of the two
structures are closely similar. Significantly, the mutant form diffracts to a
higher resolution and has a greater dimer interface. The extra dimer
interactions are located approximately 30 A away from the G526R mutation. Direct
experiments confirm the tighter dimer interaction of the G526R protein. The
results suggest the possible importance of subtle, long-range structural effects
of CMT-causing mutations at the dimer interface. From analysis of a third
crystal, an appended motif, found in higher eukaryote GlyRSs, seems not to have
a role in these long-range effects.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. G526R mutation strengthens dimer interaction. (A)
"Back" view of the GlyRS subunit that shows the dimerization
interface. This view is related to the "front" view in Fig. 1A
by a 180° rotation along the y axis. The three patches that
give dimer interactions are colored: patch 1 (F78–T137) in
cyan, patch 2 (F224–L242) in green, and patch 3 (L252–E291)
in gold. (B) Loose dimerization interface of wild-type GlyRS
generated by mapping the surface area of one subunit that is
within 7 Å of the other. (C) The same dimerization
interface generated for G526R mutant. The extra dimer interface,
absent in the wild-type enzyme, lies in the anticodon
recognition domain and is  30 Å away from the
mutation site. (D) Analytical ultracentrifugation experiment
showing that more dimers are formed by G526R mutant than by
wild-type GlyRS. (Inset) Immunoprecipitation experiment showing
that G526R mutant GlyRS pulled down more endogenous GlyRS than
did the wild-type GlyRS, presumably by forming dimers. 30 Å away from the
mutation site. (D) Analytical ultracentrifugation experiment
showing that more dimers are formed by G526R mutant than by
wild-type GlyRS. (Inset) Immunoprecipitation experiment showing
that G526R mutant GlyRS pulled down more endogenous GlyRS than
did the wild-type GlyRS, presumably by forming dimers.
|
 |
Figure 5.
Fig. 5. G41R mutation in TyrRS resembles G526R mutation in
GlyRS. (A) Active site of human TyrRS bound with substrate
analog tyrosinol. (B) CMT-causing mutation G41R would block
tyrosine binding in a similar way as G526R in GlyRS blocks
binding of the AMP moiety.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Messmer,
C.Florentz,
H.Schwenzer,
G.C.Scheper,
M.S.van der Knaap,
L.Maréchal-Drouard,
and
M.Sissler
(2011).
A human pathology-related mutation prevents import of an aminoacyl-tRNA synthetase into mitochondria.
|
| |
Biochem J,
433,
441-446.
|
 |
|
|
|
|
 |
A.Hamaguchi,
C.Ishida,
K.Iwasa,
A.Abe,
and
M.Yamada
(2010).
Charcot-Marie-Tooth disease type 2D with a novel glycyl-tRNA synthetase gene (GARS) mutation.
|
| |
J Neurol,
257,
1202-1204.
|
 |
|
|
|
|
 |
W.W.Motley,
K.Talbot,
and
K.H.Fischbeck
(2010).
GARS axonopathy: not every neuron's cup of tRNA.
|
| |
Trends Neurosci,
33,
59-66.
|
 |
|
|
|
|
 |
E.Storkebaum,
R.Leitão-Gonçalves,
T.Godenschwege,
L.Nangle,
M.Mejia,
I.Bosmans,
T.Ooms,
A.Jacobs,
P.Van Dijck,
X.L.Yang,
P.Schimmel,
K.Norga,
V.Timmerman,
P.Callaerts,
and
A.Jordanova
(2009).
Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy.
|
| |
Proc Natl Acad Sci U S A,
106,
11782-11787.
|
 |
|
|
|
|
 |
F.Achilli,
V.Bros-Facer,
H.P.Williams,
G.T.Banks,
M.AlQatari,
R.Chia,
V.Tucci,
M.Groves,
C.D.Nickols,
K.L.Seburn,
R.Kendall,
M.Z.Cader,
K.Talbot,
J.van Minnen,
R.W.Burgess,
S.Brandner,
J.E.Martin,
M.Koltzenburg,
L.Greensmith,
P.M.Nolan,
and
E.M.Fisher
(2009).
An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy.
|
| |
Dis Model Mech,
2,
359-373.
|
 |
|
|
|
|
 |
R.T.Guo,
Y.E.Chong,
M.Guo,
and
X.L.Yang
(2009).
Crystal structures and biochemical analyses suggest a unique mechanism and role for human glycyl-tRNA synthetase in Ap4A homeostasis.
|
| |
J Biol Chem,
284,
28968-28976.
|
 |
|
|
|
|
 |
A.Antonellis,
and
E.D.Green
(2008).
The role of aminoacyl-tRNA synthetases in genetic diseases.
|
| |
Annu Rev Genomics Hum Genet,
9,
87.
|
 |
|
|
|
|
 |
N.Shen,
M.Zhou,
B.Yang,
Y.Yu,
X.Dong,
and
J.Ding
(2008).
Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states.
|
| |
Nucleic Acids Res,
36,
1288-1299.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.I.Hauenstein,
Y.M.Hou,
and
J.J.Perona
(2008).
The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity.
|
| |
J Biol Chem,
283,
21997-22006.
|
 |
|
|
|
|
 |
L.A.Nangle,
W.Zhang,
W.Xie,
X.L.Yang,
and
P.Schimmel
(2007).
Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect.
|
| |
Proc Natl Acad Sci U S A,
104,
11239-11244.
|
 |
|
|
|
|
 |
X.L.Yang,
M.Kapoor,
F.J.Otero,
B.M.Slike,
H.Tsuruta,
R.Frausto,
A.Bates,
K.L.Ewalt,
D.A.Cheresh,
and
P.Schimmel
(2007).
Gain-of-function mutational activation of human tRNA synthetase procytokine.
|
| |
Chem Biol,
14,
1323-1333.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

