 |
PDBsum entry 2oyn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, unknown function
|
PDB id
|
|

|

|
2oyn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Structural genomics, unknown function
|
 |
|
Title:
|
 |
Crystal structure of cdp-bound protein mj0056 from methanococcus jannaschii, pfam duf120
|
|
Structure:
|
 |
Hypothetical protein mj0056. Chain: a. Engineered: yes
|
|
Source:
|
 |
Methanocaldococcus jannaschii dsm 2661. Organism_taxid: 243232. Strain: dsm 2661, jal-1, jcm 10045, nbrc 100440. Atcc: 43067. Gene: mj0056. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.85Å
|
R-factor:
|
0.199
|
R-free:
|
0.239
|
|
|
Authors:
|
 |
J.B.Bonanno,M.Dickey,K.T.Bain,C.Lau,R.Romero,D.Smith,S.Wasserman, J.M.Sauder,S.K.Burley,S.C.Almo,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
J.B.Bonanno
et al.
Crystal structure of hypothetical protein from methanococcus jannaschii bound to cdp.
To be published,
.
|
 |
|
Date:
|
 |
|
22-Feb-07
|
Release date:
|
06-Mar-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q60365
(RIFK_METJA) -
Riboflavin kinase from Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
132 a.a.
137 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.161
- CTP-dependent riboflavin kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
riboflavin + CTP = CDP + FMN + H+
|
 |
 |
 |
 |
 |
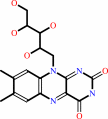 riboflavin
riboflavin
|
+
|
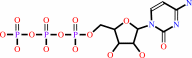 CTP
CTP
|
=
|
CDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
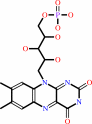 FMN
FMN
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

