 |
PDBsum entry 2ivd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2ivd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Structure of protoporphyrinogen oxidase from myxococcus xanthus with acifluorfen
|
|
Structure:
|
 |
Protoporphyrinogen oxidase. Chain: a, b. Synonym: ppo, ppox. Engineered: yes
|
|
Source:
|
 |
Myxococcus xanthus. Organism_taxid: 34. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.235
|
R-free:
|
0.283
|
|
|
Authors:
|
 |
H.R.Corradi,A.V.Corrigall,E.Boix,C.G.Mohan,E.D.Sturrock,P.N.Meissner, K.R.Acharya
|
Key ref:

|
 |
H.R.Corradi
et al.
(2006).
Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen.
J Biol Chem,
281,
38625-38633.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
12-Jun-06
|
Release date:
|
17-Oct-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P56601
(PGOX_MYXXA) -
Protoporphyrinogen oxidase from Myxococcus xanthus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
471 a.a.
449 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.3.4
- protoporphyrinogen oxidase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Porphyrin Biosynthesis (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
protoporphyrinogen IX + 3 O2 = protoporphyrin IX + 3 H2O2
|
 |
 |
 |
 |
 |
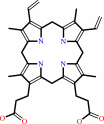 protoporphyrinogen IX
protoporphyrinogen IX
|
+
|
 3
×
O2
3
×
O2
|
=
|
 protoporphyrin IX
protoporphyrin IX
|
+
|
 3
×
H2O2
3
×
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:38625-38633
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen.
|
|
H.R.Corradi,
A.V.Corrigall,
E.Boix,
C.G.Mohan,
E.D.Sturrock,
P.N.Meissner,
K.R.Acharya.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Protoporphyrinogen IX oxidase, a monotopic membrane protein, which catalyzes the
oxidation of protoporphyrinogen IX to protoporphyrin IX in the heme/chlorophyll
biosynthetic pathway, is distributed widely throughout nature. Here we present
the structure of protoporphyrinogen IX oxidase from Myxococcus xanthus, an
enzyme with similar catalytic properties to human protoporphyrinogen IX oxidase
that also binds the common plant herbicide, acifluorfen. In the native
structure, the planar porphyrinogen substrate is mimicked by a Tween 20
molecule, tracing three sides of the macrocycle. In contrast, acifluorfen does
not mimic the planarity of the substrate but is accommodated by the shape of the
binding pocket and held in place by electrostatic and aromatic interactions. A
hydrophobic patch surrounded by positively charged residues suggests the
position of the membrane anchor, differing from the one proposed for the tobacco
mitochondrial protoporphyrinogen oxidase. Interestingly, there is a discrepancy
between the dimerization state of the protein in solution and in the crystal.
Conserved structural features are discussed in relation to a number of South
African variegate porphyria-causing mutations in the human enzyme.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. A, the structure of mxPPOX showing FAD (yellow)
and AF (green) bound and the three pseudodomains. The
pseudo-domains and some helices (H) and strands (S) are labeled.
B, overlay of mxPPOX (blue) and mtPPOX (gray). C, mxPPOX with
the residues equivalent to those involved in VP in human PPOX
displayed in pink. D, close-up of the VP equivalent residues. E,
schematic of the PPOX substrate protoporphyrinogen IX drawn with
MDL® ISIS/Draw 2.5. F, surface representation rotated
90° around the x axis from A. The membrane-binding domain
colored according to charge, showing the bottom potential
membrane-interacting surface. Blue indicates positive charge and
white/gray the hydrophobic surfaces. The FAD and
substrate-binding domains are in brown and forest green,
respectively. AF (light blue) and FAD (yellow) can be partially
seen through the hydrophobic channel.
|
 |
Figure 3.
FIGURE 3. Illustration of the crystal packing in P42[1]2;
the FAD-binding pseudo-domain is in brown, the substrate-binding
pseudo-domain in forest green, and the membrane-binding
pseudo-domain in marine blue. A, two-fold symmetry of the
asymmetric unit. B, asymmetric unit with the membrane-binding
pseudo-domain represented as a charged surface (positive charge
in blue, negative charge in red, and hydrophobic areas in gray),
showing the small area of interaction at the two-fold axis. C,
two-fold rotation of the asymmetric unit in the crystal. D,
four-fold rotation axis.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the ASBMB:
J Biol Chem
(2006,
281,
38625-38633)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.F.Hao,
Y.Tan,
N.X.Yu,
and
G.F.Yang
(2011).
Structure-activity relationships of diphenyl-ether as protoporphyrinogen oxidase inhibitors: insights from computational simulations.
|
| |
J Comput Aided Mol Des,
25,
213-222.
|
 |
|
|
|
|
 |
G.Layer,
J.Reichelt,
D.Jahn,
and
D.W.Heinz
(2010).
Structure and function of enzymes in heme biosynthesis.
|
| |
Protein Sci,
19,
1137-1161.
|
 |
|
|
|
|
 |
L.Sun,
X.Wen,
Y.Tan,
H.Li,
X.Yang,
Y.Zhao,
B.Wang,
Q.Cao,
C.Niu,
and
Z.Xi
(2009).
Site-directed mutagenesis and computational study of the Y366 active site in Bacillus subtilis protoporphyrinogen oxidase.
|
| |
Amino Acids,
37,
523-530.
|
 |
|
|
|
|
 |
L.Wang,
Y.Ma,
X.H.Liu,
Y.H.Li,
H.B.Song,
and
Z.M.Li
(2009).
Synthesis, herbicidal activities and comparative molecular field analysis study of some novel triazolinone derivatives.
|
| |
Chem Biol Drug Des,
73,
674-681.
|
 |
|
|
|
|
 |
S.Severance,
and
I.Hamza
(2009).
Trafficking of heme and porphyrins in metazoa.
|
| |
Chem Rev,
109,
4596-4616.
|
 |
|
|
|
|
 |
T.O.Boynton,
L.E.Daugherty,
T.A.Dailey,
and
H.A.Dailey
(2009).
Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity.
|
| |
Biochemistry,
48,
6705-6711.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

