 |
PDBsum entry 2e7j

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.73
- O-phospho-L-seryl-tRNA:Cys-tRNA synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-phospho-L-seryl-tRNA(Cys) + hydrogen sulfide + H+ = L-cysteinyl- tRNA(Cys) + phosphate
|
 |
 |
 |
 |
 |
O-phospho-L-seryl-tRNA(Cys)
|
+
|
hydrogen sulfide
|
+
|
H(+)
|
=
|
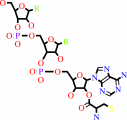 L-cysteinyl- tRNA(Cys)
L-cysteinyl- tRNA(Cys)
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
370:128-141
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus.
|
|
R.Fukunaga,
S.Yokoyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In the ancient organisms, methanogenic archaea, lacking the canonical
cysteinyl-tRNA synthetase, Cys-tRNA(Cys) is produced by an indirect pathway, in
which O-phosphoseryl-tRNA synthetase ligates O-phosphoserine (Sep) to tRNA(Cys)
and Sep-tRNA:Cys-tRNA synthase (SepCysS) converts Sep-tRNA(Cys) to
Cys-tRNA(Cys). In this study, the crystal structure of SepCysS from
Archaeoglobus fulgidus has been determined at 2.4 A resolution. SepCysS forms a
dimer, composed of monomers bearing large and small domains. The large domain
harbors the seven-stranded beta-sheet, which is typical of the pyridoxal
5'-phosphate (PLP)-dependent enzymes. In the active site, which is located near
the dimer interface, PLP is covalently bound to the side-chain of the conserved
Lys209. In the proximity of PLP, a sulfate ion is bound by the side-chains of
the conserved Arg79, His103, and Tyr104 residues. The active site is located
deep within the large, basic cleft to accommodate Sep-tRNA(Cys). On the basis of
the surface electrostatic potential, the amino acid residue conservation
mapping, the position of the bound sulfate ion, and the substrate amino acid
binding manner in other PLP-dependent enzymes, a binding model of Sep-tRNA(Cys)
to SepCysS was constructed. One of the three strictly conserved Cys residues
(Cys39, Cys42, or Cys247), of one subunit may play a crucial role in the
catalysis in the active site of the other subunit.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. The internal aldimine Lys209-PLP and the sulfate
ion in the active site. (a) Architecture of the active site
(stereo view). The main-chain traces of subunits A and B are
shown by light blue and light green tubes, respectively. The
internal aldimine Lys209-PLP (cyan) and the sulfate ion are
shown by ball and stick models. Hydrogen bonds are shown as pink
broken lines. (b) The |F[o]–F[c]| simulated-annealing omit
electron density maps (3.0σ) for Lys209-PLP and the sulfate ion
are shown in blue and green, respectively. (c) The |F[o]–F[c]|
simulated-annealing omit electron density maps (3.0σ) for
Lys209 and the sulfate ion are shown in blue and green,
respectively, in the active site of molecule B of the SeMet
SepCysS structure.
|
 |
Figure 8.
Figure 8. Active site comparison between SepCysS and CsdB and
the Sep-Ado76 binding model. (a) Architecture of the SepCysS
active site (stereo view). The main-chain traces are colored as
in Figure 7(a). The internal aldimine Lys209-PLP (cyan) and the
sulfate ion are shown by ball and stick models. (b) Architecture
of the CsdB active site (PDB ID, 1KMK) (stereo view). The
main-chain traces are colored as in Figure 7(b). The internal
aldimine Lys226-PLP and the selenocysteine are shown by cyan and
pink ball and stick models, respectively. The Cys364 side-chain
is perselenided. The coordinates of one of the oxygen atoms in
the α-COO^− group of the selenocysteine is missing in PDB ID,
1KMK. (c) Sep-Ado76 binding model. Sep-Ado76 is shown by a pink
ball and stick model. (d) Sep-Ado76 binding model on the
conservation mapping shown in Figure 6.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
370,
128-141)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Su,
M.J.Hohn,
S.Palioura,
R.L.Sherrer,
J.Yuan,
D.Söll,
and
P.O'Donoghue
(2009).
How an obscure archaeal gene inspired the discovery of selenocysteine biosynthesis in humans.
|
| |
IUBMB Life,
61,
35-39.
|
 |
|
|
|
|
 |
S.Palioura,
R.L.Sherrer,
T.A.Steitz,
D.Söll,
and
M.Simonovic
(2009).
The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation.
|
| |
Science,
325,
321-325.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.K.Bhatt,
C.Kapil,
S.Khan,
M.A.Jairajpuri,
V.Sharma,
D.Santoni,
F.Silvestrini,
E.Pizzi,
and
A.Sharma
(2009).
A genomic glimpse of aminoacyl-tRNA synthetases in malaria parasite Plasmodium falciparum.
|
| |
BMC Genomics,
10,
644.
|
 |
|
|
|
|
 |
C.M.Zhang,
C.Liu,
S.Slater,
and
Y.M.Hou
(2008).
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys).
|
| |
Nat Struct Mol Biol,
15,
507-514.
|
 |
|
|
|
|
 |
J.Yuan,
K.Sheppard,
and
D.Söll
(2008).
Amino acid modifications on tRNA.
|
| |
Acta Biochim Biophys Sin (Shanghai),
40,
539-553.
|
 |
|
|
|
|
 |
K.Sheppard,
J.Yuan,
M.J.Hohn,
B.Jester,
K.M.Devine,
and
D.Söll
(2008).
From one amino acid to another: tRNA-dependent amino acid biosynthesis.
|
| |
Nucleic Acids Res,
36,
1813-1825.
|
 |
|
|
|
|
 |
O.M.Ganichkin,
X.M.Xu,
B.A.Carlson,
H.Mix,
D.L.Hatfield,
V.N.Gladyshev,
and
M.C.Wahl
(2008).
Structure and catalytic mechanism of eukaryotic selenocysteine synthase.
|
| |
J Biol Chem,
283,
5849-5865.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.I.Hauenstein,
and
J.J.Perona
(2008).
Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei.
|
| |
J Biol Chem,
283,
22007-22017.
|
 |
|
|
|
|
 |
Y.Araiso,
S.Palioura,
R.Ishitani,
R.L.Sherrer,
P.O'Donoghue,
J.Yuan,
H.Oshikane,
N.Domae,
J.Defranco,
D.Söll,
and
O.Nureki
(2008).
Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation.
|
| |
Nucleic Acids Res,
36,
1187-1199.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

