 |
PDBsum entry 2dtv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
 |
Obsolete entry |
 |
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of lysn, alpha-aminoadipate aminotransferase, from thermus thermophilus hb27
|
|
Structure:
|
 |
Alpha-aminodipate aminotransferase. Chain: a, b, c, d, e, f. Engineered: yes
|
|
Source:
|
 |
Thermus thermophilus. Bacteria. Strain: hb27. Gene: lysn. Expressed in: escherichia coli.
|
|
Resolution:
|
 |
|
2.26Å
|
R-factor:
|
0.220
|
R-free:
|
0.269
|
|
|
Authors:
|
 |
T.Tomita,T.Miyazaki,T.Miyagawa,S.Fushinobu,T.Kuzuyama, M.Nishiyama
|
|
Key ref:
|
 |
T.Tomita
et al.
Crystal structure of alpha-Aminoadipate aminotransferase, A homolog of human kynurenine aminotransferase ii, From thermus thermophilus.
To be published,
.
|
 |
|
Date:
|
 |
|
17-Jul-06
|
Release date:
|
17-Jul-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q72LL6
(LYSN_THET2) -
2-aminoadipate transaminase from Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
397 a.a.
391 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.39
- 2-aminoadipate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Lysine catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-2-aminoadipate + 2-oxoglutarate = 2-oxoadipate + L-glutamate
|
 |
 |
 |
 |
 |
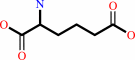 L-2-aminoadipate
L-2-aminoadipate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 2-oxoadipate
2-oxoadipate
|
+
|
L-glutamate
Bound ligand (Het Group name = )
matches with 72.73% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

