 |
PDBsum entry 2c4e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.7.1.213
- cytidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cytidine + ATP = CMP + ADP + H+
|
 |
 |
 |
 |
 |
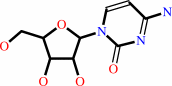 cytidine
cytidine
|
+
|
 ATP
ATP
|
=
|
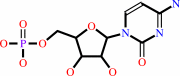 CMP
CMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.1.73
- inosine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
inosine + ATP = IMP + ADP + H+
|
 |
 |
 |
 |
 |
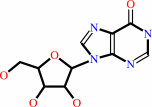 inosine
inosine
|
+
|
 ATP
ATP
|
=
|
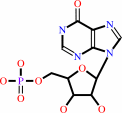 IMP
IMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
62:1085-1097
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of Methanocaldococcus jannaschii nucleoside kinase: an archaeal member of the ribokinase family.
|
|
L.Arnfors,
T.Hansen,
P.Schönheit,
R.Ladenstein,
W.Meining.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nucleoside kinase from the hyperthermophilic archaeon Methanocaldococcus
jannaschii (MjNK) is a member of the ribokinase family. In the presence of ATP
and Mg(2+), MjNK is able to catalyze the phosphorylation of a variety of
nucleosides, including inosine, cytidine, guanosine and adenosine. Here, the
crystal structure of MjNK, the first structure of an archaeal representative of
the ribokinase family, is presented. The structure was solved using the
multiple-wavelength anomalous dispersion technique. Three-dimensional structures
of the unliganded enzyme and a complex of MjNK, an ATP analogue and adenosine
were determined to 1.7 and 1.9 A resolution, respectively. Each subunit
comprises an alpha/beta-domain and a smaller lid domain and has an overall fold
characteristic of the ribokinase superfamily. MjNK shares highest structural
similarity to the ribokinases from Escherichia coli and Thermotoga maritima.
Similar to ribokinase and other superfamily members, the lid domain of MjNK
undergoes a significant conformational change upon substrate binding. In the
crystal structure of the MjNK complex, subunit A adopts a closed conformation
and subunit B an open conformation. In subunit A all substrates and Mg(2+) were
observed, whereas in subunit B only the ATP analogue could be clearly identified
in the electron density. The structures of MjNK and E. coli ribokinase (EcRK)
were compared with respect to putative determinants of thermal stability.
Relative to EcRK, MjNK shows an increased charged and a decreased hydrophobic
accessible surface area, as well as a higher fraction of charged residues, ionic
networks and large aromatic clusters, characteristics that are frequently
observed in enzymes from hyperthermophiles.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Ribbon representation of MjNK. (a) Crystallographic
dimer of the apo form (subunit A, purple; subunit A', pink) and
(b) in complex with adenosine (red) and AMP-PNP (yellow).
Subunit A (dark blue) is in the closed conformation with the lid
domain closer to the  / /
 -domain
and contains the strongly bound adenosine, whereas subunit B
(light blue) is in the open conformation and contains an
adenosine that is bound with low occupancy. Mg atoms and
surrounding waters are shown as green and red spheres,
respectively. -domain
and contains the strongly bound adenosine, whereas subunit B
(light blue) is in the open conformation and contains an
adenosine that is bound with low occupancy. Mg atoms and
surrounding waters are shown as green and red spheres,
respectively.
|
 |
Figure 7.
Figure 7 Schematic overview of contacts between adenosine
(red), AMP-PNP (green), the solvated magnesium ion (blue) and
the protein environment (black). Dashed lines indicate atomic
distances between non-C atoms of less than 3.5 Å. The
imidazole ring of adenosine is sandwiched between the aromatic
ring of Phe113 and the amid group of Gln163.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
1085-1097)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Ota,
S.Sakasegawa,
Y.Yasuda,
S.Imamura,
and
T.Tamura
(2008).
A novel nucleoside kinase from Burkholderia thailandensis.
|
| |
FEBS J,
275,
5865-5872.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

