 |
PDBsum entry 1xpq

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1xpq
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of fms1, a polyamine oxidase from yeast
|
|
Structure:
|
 |
Polyamine oxidase fms1. Chain: a, b, c, d. Synonym: fenpropimorph resistance multicopy suppressor 1. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Gene: fms1, ymr020w, ym9711.09. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.51Å
|
R-factor:
|
0.202
|
R-free:
|
0.276
|
|
|
Authors:
|
 |
Q.Huang,Q.Liu,Q.Hao
|
Key ref:

|
 |
Q.Huang
et al.
(2005).
Crystal structures of Fms1 and its complex with spermine reveal substrate specificity.
J Mol Biol,
348,
951-959.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Oct-04
|
Release date:
|
26-Apr-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P50264
(FMS1_YEAST) -
Polyamine oxidase FMS1 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
508 a.a.
502 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.5.3.17
- non-specific polyamine oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
spermine + O2 + H2O = 3-aminopropanal + spermidine + H2O2
|
|
2.
|
spermidine + O2 + H2O = 3-aminopropanal + putrescine + H2O2
|
|
3.
|
N1-acetylspermine + O2 + H2O = 3-acetamidopropanal + spermidine + H2O2
|
|
4.
|
N1-acetylspermidine + O2 + H2O = 3-acetamidopropanal + putrescine + H2O2
|
|
 |
 |
 |
 |
 |
spermine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
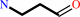 3-aminopropanal
3-aminopropanal
|
+
|
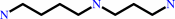 spermidine
spermidine
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
spermidine
Bound ligand (Het Group name = )
matches with 71.43% similarity
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
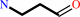 3-aminopropanal
3-aminopropanal
|
+
|
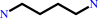 putrescine
putrescine
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 N(1)-acetylspermine
N(1)-acetylspermine
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
3-acetamidopropanal
Bound ligand (Het Group name = )
matches with 71.43% similarity
|
+
|
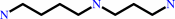 spermidine
spermidine
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 N(1)-acetylspermidine
N(1)-acetylspermidine
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
3-acetamidopropanal
Bound ligand (Het Group name = )
matches with 42.86% similarity
|
+
|
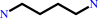 putrescine
putrescine
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
348:951-959
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of Fms1 and its complex with spermine reveal substrate specificity.
|
|
Q.Huang,
Q.Liu,
Q.Hao.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Fms1 is a rate-limiting enzyme for the biosynthesis of pantothenic acid in
yeast. Fms1 has polyamine oxidase (PAO) activity, which converts spermine into
spermidine and 3-aminopropanal. The 3-aminopropanal is further oxidized to
produce beta-alanine, which is necessary for the biosynthesis of pantothenic
acid. The crystal structures of Fms1 and its complex with the substrate spermine
have been determined using the single-wavelength anomalous diffraction (SAD)
phasing method. Fms1 consists of an FAD-binding domain, with Rossmann fold
topology, and a substrate-binding domain. The active site is a tunnel located at
the interface of the two domains. The substrate spermine binds to the active
site mainly via hydrogen bonds and hydrophobic interactions. In the complex, C11
but not C9 of spermine is close enough to the catalytic site (N5 of FAD) to be
oxidized. Therefore, the products are spermidine and 3-aminopropanal, rather
than 3-(aminopropyl) 4-aminobutyraldehyde and 1,3-diaminoprone.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. (a) The F[o]-F[c] omit map contoured at 4s
showing spermine in the complex. (b) In the complex, the
substrate spermine binds to the active site of the enzyme mainly
through two hydrogen bonds and hydrophobic interactions,
especially the W174-spermine (C6-C9)-W65 hydrophobic sandwich.
The C11 atom is close enough (3.76 Å) to the N5 atom of
FAD to be oxidized, while C9 is too far from N5 of FAD to be
oxidized. (c) A LIGPLOT42 outlining the substrate-tunnel
interactions.
|
 |
Figure 4.
Figure 4. The binding of the substrate spermine to the
active site of Fms1 induces conformation changes of the protein.
The side-chain of W174 (native in green; complex in red) moves
to a new position to form the W174-spermine-W65 hydrophobic
sandwich.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
348,
951-959)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Fiorillo,
R.Federico,
F.Polticelli,
A.Boffi,
F.Mazzei,
M.Di Fusco,
A.Ilari,
and
P.Tavladoraki
(2011).
The structure of maize polyamine oxidase K300M mutant in complex with the natural substrates provides a snapshot of the catalytic mechanism of polyamine oxidation.
|
| |
FEBS J,
278,
809-821.
|
 |
|
|
|
|
 |
P.Tavladoraki,
M.Cervelli,
F.Antonangeli,
G.Minervini,
P.Stano,
R.Federico,
P.Mariottini,
and
F.Polticelli
(2011).
Probing mammalian spermine oxidase enzyme-substrate complex through molecular modeling, site-directed mutagenesis and biochemical characterization.
|
| |
Amino Acids,
40,
1115-1126.
|
 |
|
|
|
|
 |
M.Cervelli,
G.Bellavia,
E.Fratini,
R.Amendola,
F.Polticelli,
M.Barba,
R.Federico,
F.Signore,
G.Gucciardo,
R.Grillo,
P.M.Woster,
R.A.Casero,
and
P.Mariottini
(2010).
Spermine oxidase (SMO) activity in breast tumor tissues and biochemical analysis of the anticancer spermine analogues BENSpm and CPENSpm.
|
| |
BMC Cancer,
10,
555.
|
 |
|
|
|
|
 |
M.S.Adachi,
P.R.Juarez,
and
P.F.Fitzpatrick
(2010).
Mechanistic studies of human spermine oxidase: kinetic mechanism and pH effects.
|
| |
Biochemistry,
49,
386-392.
|
 |
|
|
|
|
 |
P.F.Fitzpatrick
(2010).
Oxidation of amines by flavoproteins.
|
| |
Arch Biochem Biophys,
493,
13-25.
|
 |
|
|
|
|
 |
F.Forneris,
E.Battaglioli,
A.Mattevi,
and
C.Binda
(2009).
New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin.
|
| |
FEBS J,
276,
4304-4312.
|
 |
|
|
|
|
 |
H.Gaweska,
M.Henderson Pozzi,
D.M.Schmidt,
D.G.McCafferty,
and
P.F.Fitzpatrick
(2009).
Use of pH and kinetic isotope effects to establish chemistry as rate-limiting in oxidation of a peptide substrate by LSD1.
|
| |
Biochemistry,
48,
5440-5445.
|
 |
|
|
|
|
 |
M.H.Pozzi,
V.Gawandi,
and
P.F.Fitzpatrick
(2009).
Mechanistic studies of para-substituted N,N'-dibenzyl-1,4-diaminobutanes as substrates for a mammalian polyamine oxidase.
|
| |
Biochemistry,
48,
12305-12313.
|
 |
|
|
|
|
 |
M.Henderson Pozzi,
V.Gawandi,
and
P.F.Fitzpatrick
(2009).
pH dependence of a mammalian polyamine oxidase: insights into substrate specificity and the role of lysine 315.
|
| |
Biochemistry,
48,
1508-1516.
|
 |
|
|
|
|
 |
K.Ida,
M.Kurabayashi,
M.Suguro,
Y.Hiruma,
T.Hikima,
M.Yamomoto,
and
H.Suzuki
(2008).
Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
|
| |
J Biol Chem,
283,
16584-16590.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Webb,
A.Marquet,
R.R.Mendel,
F.Rébeillé,
and
A.G.Smith
(2007).
Elucidating biosynthetic pathways for vitamins and cofactors.
|
| |
Nat Prod Rep,
24,
988.
|
 |
|
|
|
|
 |
P.F.Fitzpatrick
(2007).
Insights into the mechanisms of flavoprotein oxidases from kinetic isotope effects.
|
| |
J Labelled Comp Radiopharm,
50,
1016-1025.
|
 |
|
|
|
|
 |
M.Bianchi,
F.Polticelli,
P.Ascenzi,
M.Botta,
R.Federico,
P.Mariottini,
and
A.Cona
(2006).
Inhibition of polyamine and spermine oxidases by polyamine analogues.
|
| |
FEBS J,
273,
1115-1123.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

