 |
PDBsum entry 1wmb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1wmb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.30
- 3-hydroxybutyrate dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-3-hydroxybutanoate + NAD+ = acetoacetate + NADH + H+
|
 |
 |
 |
 |
 |
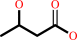 (R)-3-hydroxybutanoate
(R)-3-hydroxybutanoate
|
+
|
 NAD(+)
NAD(+)
|
=
|
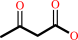 acetoacetate
acetoacetate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
355:722-733
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
D-3-hydroxybutyrate dehydrogenase from Pseudomonas fragi: molecular cloning of the enzyme gene and crystal structure of the enzyme.
|
|
K.Ito,
Y.Nakajima,
E.Ichihara,
K.Ogawa,
N.Katayama,
K.Nakashima,
T.Yoshimoto.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The gene coding for d-3-hydroxybutyrate dehydrogenase (HBDH) was cloned from
Pseudomonas fragi. The nucleotide sequence contained a 780 bp open reading frame
encoding a 260 amino acid residue protein. The recombinant enzyme was
efficiently expressed in Escherichia coli cells harboring pHBDH11 and was
purified to homogeneity as judged by SDS-PAGE. The enzyme showed a strict
stereospecificity to the D-enantiomer (3R-configuration) of 3-hydroxybutyrate as
a substrate. Crystals of the ligand-free HBDH and of the enzyme-NAD+ complex
were obtained using the hanging-drop, vapor-diffusion method. The crystal
structure of the HBDH was solved by the multiwavelength anomalous diffraction
method using the SeMet-substituted enzyme and was refined to 2.0 A resolution.
The overall structure of P.fragi HBDH, including the catalytic tetrad of Asn114,
Ser142, Tyr155, and Lys159, shows obvious relationships with other members of
the short-chain dehydrogenase/reductase (SDR) family. A cacodylate anion was
observed in both the ligand-free enzyme and the enzyme-NAD+ complex, and was
located near the catalytic tetrad. It was shown that the cacodylate inhibited
the NAD+-dependent D-3-hydroxybutyrate dehydrogenation competitively, with a Ki
value of 5.6 mM. From the interactions between cacodylate and the enzyme, it is
predicted that substrate specificity is achieved through the recognition of the
3-methyl and carboxyl groups of the substrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. The stereo diagram of SDRs superimposed onto
ligand-free HBDH. The C^a trace of HBDH is shown by the orange
line, and 2hsd, 1ae1, 1k2w, 1gco, 1hxh, and 1ahh by red, blue,
cyan, blue-violet, magenta, and green lines, respectively.
|
 |
Figure 5.
Figure 5. Drawing of the NAD^+ binding site in HBDH. (a)
The wire frame shows the F[o] -F[c] omit map for NAD^+ and
cacodylate anion contoured at 2s levels. The stick models show
residues interacting with NAD^+ or cacodylate anion. (b) The
scheme diagram shows hydrogen-bonding interaction with NAD^+.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2006,
355,
722-733)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Mountassif,
P.Andreoletti,
M.Cherkaoui-Malki,
N.Latruffe,
and
M.S.El Kebbaj
(2010).
Structural and catalytic properties of the D-3-hydroxybutyrate dehydrogenase from Pseudomonas aeruginosa.
|
| |
Curr Microbiol,
61,
7.
|
 |
|
|
|
|
 |
F.H.Niesen,
L.Schultz,
A.Jadhav,
C.Bhatia,
K.Guo,
D.J.Maloney,
E.S.Pilka,
M.Wang,
U.Oppermann,
T.D.Heightman,
and
A.Simeonov
(2010).
High-affinity inhibitors of human NAD-dependent 15-hydroxyprostaglandin dehydrogenase: mechanisms of inhibition and structure-activity relationships.
|
| |
PLoS One,
5,
e13719.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Nakashima,
K.Ito,
Y.Nakajima,
R.Yamazawa,
S.Miyakawa,
and
T.Yoshimoto
(2009).
Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
|
| |
J Biochem,
145,
467-479.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.M.Hoque,
S.Shimizu,
E.C.Juan,
Y.Sato,
M.T.Hossain,
T.Yamamoto,
S.Imamura,
K.Suzuki,
H.Amano,
T.Sekiguchi,
M.Tsunoda,
and
A.Takénaka
(2009).
Structure of D-3-hydroxybutyrate dehydrogenase prepared in the presence of the substrate D-3-hydroxybutyrate and NAD+.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
331-335.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Mountassif,
P.Andreoletti,
Z.El Kebbaj,
A.Moutaouakkil,
M.Cherkaoui-Malki,
N.Latruffe,
and
M.S.El Kebbaj
(2008).
Immunoaffinity purification and characterization of mitochondrial membrane-bound D-3-hydroxybutyrate dehydrogenase from Jaculus orientalis.
|
| |
BMC Biochem,
9,
26.
|
 |
|
|
|
|
 |
M.M.Hoque,
S.Shimizu,
M.T.Hossain,
T.Yamamoto,
S.Imamura,
K.Suzuki,
M.Tsunoda,
H.Amano,
T.Sekiguchi,
and
A.Takénaka
(2008).
The structures of Alcaligenes faecalis D-3-hydroxybutyrate dehydrogenase before and after NAD+ and acetate binding suggest a dynamical reaction mechanism as a member of the SDR family.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
496-505.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.S.Paithankar,
C.Feller,
E.B.Kuettner,
A.Keim,
M.Grunow,
and
N.Sträter
(2007).
Cosubstrate-induced dynamics of D-3-hydroxybutyrate dehydrogenase from Pseudomonas putida.
|
| |
FEBS J,
274,
5767-5779.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Yoshimoto
(2007).
[Biochemistry and structural biology of microbial enzymes and their medical applications]
|
| |
Yakugaku Zasshi,
127,
1035-1045.
|
 |
|
|
|
|
 |
C.Feller,
R.Günther,
H.J.Hofmann,
and
M.Grunow
(2006).
Molecular basis of substrate recognition in D-3-hydroxybutyrate dehydrogenase from Pseudomonas putida.
|
| |
Chembiochem,
7,
1410-1418.
|
 |
|
|
|
|
 |
S.Nakamura,
M.Oda,
S.Kataoka,
S.Ueda,
S.Uchiyama,
T.Yoshida,
Y.Kobayashi,
and
T.Ohkubo
(2006).
Apo- and holo-structures of 3alpha-hydroxysteroid dehydrogenase from Pseudomonas sp. B-0831. Loop-helix transition induced by coenzyme binding.
|
| |
J Biol Chem,
281,
31876-31884.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

