 |
PDBsum entry 1wkv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of o-phosphoserine sulfhydrylase
|
|
Structure:
|
 |
Cysteine synthase. Chain: a, b. Synonym: o-phosphoserine sulfhydrylase. Engineered: yes
|
|
Source:
|
 |
Aeropyrum pernix. Organism_taxid: 272557. Strain: k1. Gene: ape1586. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.190
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
Y.Oda,K.Mino,K.Ishikawa,M.Ataka
|
Key ref:

|
 |
Y.Oda
et al.
(2005).
Three-dimensional structure of a new enzyme, O-phosphoserine sulfhydrylase, involved in l-cysteine biosynthesis by a hyperthermophilic archaeon, Aeropyrum pernix K1, at 2.0A resolution.
J Mol Biol,
351,
334-344.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Jun-04
|
Release date:
|
28-Jun-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9YBL2
(CYSO_AERPE) -
Protein CysO from Aeropyrum pernix (strain ATCC 700893 / DSM 11879 / JCM 9820 / NBRC 100138 / K1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
389 a.a.
382 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.47
- cysteine synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-acetyl-L-serine + hydrogen sulfide = L-cysteine + acetate
|
 |
 |
 |
 |
 |
 O-acetyl-L-serine
O-acetyl-L-serine
|
+
|
hydrogen sulfide
|
=
|
 L-cysteine
L-cysteine
|
+
|
acetate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.5.1.65
- O-phosphoserine sulfhydrylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-phospho-L-serine + hydrogen sulfide + H+ = L-cysteine + phosphate
|
 |
 |
 |
 |
 |
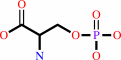 O-phospho-L-serine
O-phospho-L-serine
|
+
|
hydrogen sulfide
|
+
|
H(+)
|
=
|
L-cysteine
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.4.2.1.22
- cystathionine beta-synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-homocysteine + L-serine = L,L-cystathionine + H2O
|
 |
 |
 |
 |
 |
 L-homocysteine
L-homocysteine
|
+
|
L-serine
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
=
|
L,L-cystathionine
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
351:334-344
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of a new enzyme, O-phosphoserine sulfhydrylase, involved in l-cysteine biosynthesis by a hyperthermophilic archaeon, Aeropyrum pernix K1, at 2.0A resolution.
|
|
Y.Oda,
K.Mino,
K.Ishikawa,
M.Ataka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
O-Phosphoserine sulfhydrylase is a new enzyme found in a hyperthermophilic
archaeon, Aeropyrum pernix K1. This enzyme catalyzes a novel cysteine synthetic
reaction from O-phospho-l-serine and sulfide. The crystal structure of the
enzyme was determined at 2.0A resolution using the method of multi-wavelength
anomalous dispersion. A monomer consists of three domains, including an
N-terminal domain with a new alpha/beta fold. The topology folds of the middle
and C-terminal domains were similar to those of the O-acetylserine
sulfhydrylase-A from Salmonella typhimurium and the cystathionine beta-synthase
from human. The cofactor, pyridoxal 5'-phosphate, is bound in a cleft between
the middle and C-terminal domains through a covalent linkage to Lys127. Based on
the structure determined, O-phospho-l-serine could be rationally modeled into
the active site of the enzyme. An enzyme-substrate complex model and a mutation
experiment revealed that Arg297, unique to hyperthermophilic archaea, is one of
the most crucial residues for O-phosphoserine sulfhydrylation activity. There
are more hydrophobic areas and less electric charges at the dimer interface,
compared to the S.typhimurium O-acetylserine sulfhydrylase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. A detailed structure of the active site of OPSS.
(a) Stereo view of the active site region of OPSS. The cofactor
PLP and the bound acetate ion are located at the center of the
active site. Their 2F[o] -F[c] electron density is shown at 2s.
Hydrogen bonds are shown as broken lines. A Schiff base linkage
is shown as a thick, broken blue line. (b) A schematic of
hydrogen bond interactions among PLP moiety, protein and water
molecules. (c) Stereo view of a proposed model of the OPSS
active site with phosphoserine. Phosphoserine was built into the
active site to make a Schiff base linkage with PLP and to bind
with Thr152, Ser153, and Gln224. The negative charge of the
phosphate group of phosphoserine is presumably interacting with
Arg297. The Figure was produced with MOLSCRIPT.29
|
 |
Figure 4.
Figure 4. Stereo views of comparisons of the active site
regions between OPSS and S. typhimurium OASS or human CBS. (a) A
superposition of OPSS (green) and the mutant OASS bound with
methionine (gray). (b) A superposition of OPSS (green) and the
free form of wild-type OASS (gray). (c) A superposition of OPSS
(green) and CBS (gray). In CBS, a residue corresponding to
Thr203 of OPSS is not visible in its crystallographic structure.
The Figure was produced with MOLSCRIPT29 and Raster3D.30
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
351,
334-344)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.E.Graham,
S.M.Taylor,
R.Z.Wolf,
and
S.C.Namboori
(2009).
Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase.
|
| |
Biochem J,
424,
467-478.
|
 |
|
|
|
|
 |
R.A.Williams,
G.D.Westrop,
and
G.H.Coombs
(2009).
Two pathways for cysteine biosynthesis in Leishmania major.
|
| |
Biochem J,
420,
451-462.
|
 |
|
|
|
|
 |
D.Agren,
R.Schnell,
W.Oehlmann,
M.Singh,
and
G.Schneider
(2008).
Cysteine Synthase (CysM) of Mycobacterium tuberculosis Is an O-Phosphoserine Sulfhydrylase: EVIDENCE FOR AN ALTERNATIVE CYSTEINE BIOSYNTHESIS PATHWAY IN MYCOBACTERIA.
|
| |
J Biol Chem,
283,
31567-31574.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Anderson,
J.Rodriguez,
D.Susanti,
I.Porat,
C.Reich,
L.E.Ulrich,
J.G.Elkins,
K.Mavromatis,
A.Lykidis,
E.Kim,
L.S.Thompson,
M.Nolan,
M.Land,
A.Copeland,
A.Lapidus,
S.Lucas,
C.Detter,
I.B.Zhulin,
G.J.Olsen,
W.Whitman,
B.Mukhopadhyay,
J.Bristow,
and
N.Kyrpides
(2008).
Genome sequence of Thermofilum pendens reveals an exceptional loss of biosynthetic pathways without genome reduction.
|
| |
J Bacteriol,
190,
2957-2965.
|
 |
|
|
|
|
 |
L.Klipcan,
M.Frenkel-Morgenstern,
and
M.G.Safro
(2008).
Presence of tRNA-dependent pathways correlates with high cysteine content in methanogenic Archaea.
|
| |
Trends Genet,
24,
59-63.
|
 |
|
|
|
|
 |
S.I.Hauenstein,
and
J.J.Perona
(2008).
Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei.
|
| |
J Biol Chem,
283,
22007-22017.
|
 |
|
|
|
|
 |
G.D.Westrop,
G.Goodall,
J.C.Mottram,
and
G.H.Coombs
(2006).
Cysteine biosynthesis in Trichomonas vaginalis involves cysteine synthase utilizing O-phosphoserine.
|
| |
J Biol Chem,
281,
25062-25075.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

