 |
PDBsum entry 1tpc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Isomerase(intramolecular oxidoreductase)
|
PDB id
|
|

|

|
1tpc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.2.3.3
- methylglyoxal synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dihydroxyacetone phosphate = methylglyoxal + phosphate
|
 |
 |
 |
 |
 |
dihydroxyacetone phosphate
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
=
|
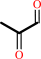 methylglyoxal
methylglyoxal
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.3.1.1
- triose-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glyceraldehyde 3-phosphate = dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
D-glyceraldehyde 3-phosphate
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
=
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
34:13612-13621
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
|
|
E.A.Komives,
J.C.Lougheed,
K.Liu,
S.Sugio,
Z.Zhang,
G.A.Petsko,
D.Ringe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structural basis for the improvement in catalytic efficiency of the mutant
E165D chicken triosephosphate isomerase by the secondary mutation, S96P, has
been analyzed using a combination of X-ray crystallography and Fourier transform
infrared spectroscopy. All X-ray structures were of the complex of
phosphoglycolohydroxamate (PGH), an intermediate analog, with the isomerase, and
each was solved to a resolution of 1.9 A. Comparison of the structure of the
double mutant, E165D.S96P, with that of the single mutant, E165D, as well as
with the wild-type isomerase shows only insignificant differences in the
positions of the side chains in all of the mutants when compared with the
wild-type isomerase, except that in both the E165D and E165D.S96P mutants, the
aspartate side chain was approximately 0.7 A further away from the substrate
analog than the glutamate side chain. Significant differences were observed in
the crystal structure of the E165D.S96P double mutant in the positions of
ordered water molecules bound at the active site. The loss of two water
molecules located near the side chain at position 165 was observed in isomerases
containing the S96P mutation. The resulting increase in hydrophobicity of the
pocket probably causes an increase in the pKa of the catalytic base, D165,
thereby improving its basicity. A new ordered water molecule was observed
underneath the bound PGH in the E165D.S96P structure, which likely decreases the
pKa's of the substrate protons, thereby increasing their acidity. An enzyme
derived carbonyl stretch at 1746 cm-1 that is only observed in the IR spectrum
of the E165D.S96P double mutant isomerase with bound substrates has been
assigned to a stable ground state protonated D165-enediol(ate) intermediate
complex. Thus, the gain in activity resulting from the S96P second site change
probably results from a combination of improving the basicity of the enzyme,
improving the acidity of the substrate protons, and stabilization of a reaction
intermediate. All three of these effects seem to be caused by changes in bound
water molecules.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Jogl,
S.Rozovsky,
A.E.McDermott,
and
L.Tong
(2003).
Optimal alignment for enzymatic proton transfer: structure of the Michaelis complex of triosephosphate isomerase at 1.2-A resolution.
|
| |
Proc Natl Acad Sci U S A,
100,
50-55.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Ringe,
and
C.Mattos
(1999).
Analysis of the binding surfaces of proteins.
|
| |
Med Res Rev,
19,
321-331.
|
 |
|
|
|
|
 |
S.Shaltiel,
S.Cox,
and
S.S.Taylor
(1998).
Conserved water molecules contribute to the extensive network of interactions at the active site of protein kinase A.
|
| |
Proc Natl Acad Sci U S A,
95,
484-491.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

