 |
PDBsum entry 1sg0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1sg0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure analysis of qr2 in complex with resveratrol
|
|
Structure:
|
 |
Nrh dehydrogenase [quinone] 2. Chain: a, b. Synonym: quinone reductase 2, qr2, nrh:quinone oxidoreductase 2. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: nqo2, nmor2, bc006096. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
1.50Å
|
R-factor:
|
0.214
|
R-free:
|
0.234
|
|
|
Authors:
|
 |
L.Buryanovskyy,Y.Fu,M.Boyd,Y.Ma,T.C.Tsieh,J.M.Wu,Z.Zhang
|
Key ref:

|
 |
L.Buryanovskyy
et al.
(2004).
Crystal structure of quinone reductase 2 in complex with resveratrol.
Biochemistry,
43,
11417-11426.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
22-Feb-04
|
Release date:
|
25-Jan-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P16083
(NQO2_HUMAN) -
Ribosyldihydronicotinamide dehydrogenase [quinone] from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
231 a.a.
230 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.10.5.1
- ribosyldihydronicotinamide dehydrogenase (quinone).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide + a quinone + H+ = beta-nicotinamide D-riboside + a quinol
|
 |
 |
 |
 |
 |
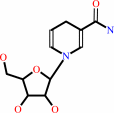 1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide
1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide
|
+
|
 quinone
quinone
|
+
|
H(+)
|
=
|
beta-nicotinamide D-riboside
|
+
|
quinol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Zn(2+)
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
Zn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:11417-11426
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of quinone reductase 2 in complex with resveratrol.
|
|
L.Buryanovskyy,
Y.Fu,
M.Boyd,
Y.Ma,
T.C.Hsieh,
J.M.Wu,
Z.Zhang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Resveratrol has been shown to have chemopreventive, cardioprotective, and
antiaging properties. Here, we report that resveratrol is a potent inhibitor of
quinone reductase 2 (QR2) activity in vitro with a dissociation constant of 35
nM and show that it specifically binds to the deep active-site cleft of QR2
using high-resolution structural analysis. All three resveratrol hydroxyl groups
form hydrogen bonds with amino acids from QR2, anchoring a flat resveratrol
molecule in parallel with the isoalloxazine ring of FAD. The unique active-site
pocket in QR2 could potentially bind other natural polyphenols such as
flavonoids, as proven by the high affinity exhibited by quercetin toward QR2.
K562 cells with QR2 expression suppressed by RNAi showed similar properties as
resveratrol-treated cells in their resistance to quinone toxicity. Furthermore,
the QR2 knockdown K562 cells exhibit increased antioxidant and detoxification
enzyme expression and reduced proliferation rates. These observations could
imply that the chemopreventive and cardioprotective properties of resveratrol
are possibly the results of QR2 activity inhibition, which in turn, up-regulates
the expression of cellular antioxidant enzymes and cellular resistance to
oxidative stress.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.F.Ruan,
X.Lu,
J.F.Tang,
Y.Wei,
X.L.Wang,
Y.B.Zhang,
L.S.Wang,
and
H.L.Zhu
(2011).
Synthesis, biological evaluation, and molecular docking studies of resveratrol derivatives possessing chalcone moiety as potential antitubulin agents.
|
| |
Bioorg Med Chem,
19,
2688-2695.
|
 |
|
|
|
|
 |
J.M.Pezzuto
(2011).
The phenomenon of resveratrol: redefining the virtues of promiscuity.
|
| |
Ann N Y Acad Sci,
1215,
123-130.
|
 |
|
|
|
|
 |
J.M.Wu,
and
T.C.Hsieh
(2011).
Resveratrol: a cardioprotective substance.
|
| |
Ann N Y Acad Sci,
1215,
16-21.
|
 |
|
|
|
|
 |
A.A.Khutornenko,
V.V.Roudko,
B.V.Chernyak,
A.B.Vartapetian,
P.M.Chumakov,
and
A.G.Evstafieva
(2010).
Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway.
|
| |
Proc Natl Acad Sci U S A,
107,
12828-12833.
|
 |
|
|
|
|
 |
B.Calamini,
K.Ratia,
M.G.Malkowski,
M.Cuendet,
J.M.Pezzuto,
B.D.Santarsiero,
and
A.D.Mesecar
(2010).
Pleiotropic mechanisms facilitated by resveratrol and its metabolites.
|
| |
Biochem J,
429,
273-282.
|
 |
|
|
|
|
 |
G.F.Oxenkrug,
S.O.Bachurin,
I.V.Prakhie,
and
N.S.Zefirov
(2010).
Quinone reductase 2 and antidepressant effect of melatonin derivatives.
|
| |
Ann N Y Acad Sci,
1199,
121-124.
|
 |
|
|
|
|
 |
J.A.Baur
(2010).
Resveratrol, sirtuins, and the promise of a DR mimetic.
|
| |
Mech Ageing Dev,
131,
261-269.
|
 |
|
|
|
|
 |
J.H.Yang,
T.P.Kondratyuk,
L.E.Marler,
X.Qiu,
Y.Choi,
H.Cao,
R.Yu,
M.Sturdy,
S.Pegan,
Y.Liu,
L.Q.Wang,
A.D.Mesecar,
R.B.Van Breemen,
J.M.Pezzuto,
H.H.Fong,
Y.G.Chen,
and
H.J.Zhang
(2010).
Isolation and evaluation of kaempferol glycosides from the fern Neocheiropteris palmatopedata.
|
| |
Phytochemistry,
71,
641-647.
|
 |
|
|
|
|
 |
R.H.Singleton,
H.Q.Yan,
W.Fellows-Mayle,
and
C.E.Dixon
(2010).
Resveratrol attenuates behavioral impairments and reduces cortical and hippocampal loss in a rat controlled cortical impact model of traumatic brain injury.
|
| |
J Neurotrauma,
27,
1091-1099.
|
 |
|
|
|
|
 |
T.C.Hsieh,
and
J.M.Wu
(2010).
Resveratrol: Biological and pharmaceutical properties as anticancer molecule.
|
| |
Biofactors,
36,
360-369.
|
 |
|
|
|
|
 |
A.Maiti,
P.V.Reddy,
M.Sturdy,
L.Marler,
S.D.Pegan,
A.D.Mesecar,
J.M.Pezzuto,
and
M.Cushman
(2009).
Synthesis of casimiroin and optimization of its quinone reductase 2 and aromatase inhibitory activities.
|
| |
J Med Chem,
52,
1873-1884.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Winger,
O.Hantschel,
G.Superti-Furga,
and
J.Kuriyan
(2009).
The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2).
|
| |
BMC Struct Biol,
9,
7.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.M.Pezzuto,
V.Venkatasubramanian,
M.Hamad,
and
K.R.Morris
(2009).
Unraveling the relationship between grapes and health.
|
| |
J Nutr,
139,
1783S-1787S.
|
 |
|
|
|
|
 |
S.Pervaiz,
and
A.L.Holme
(2009).
Resveratrol: its biologic targets and functional activity.
|
| |
Antioxid Redox Signal,
11,
2851-2897.
|
 |
|
|
|
|
 |
C.Yan,
J.K.Kepa,
D.Siegel,
I.J.Stratford,
and
D.Ross
(2008).
Dissecting the role of multiple reductases in bioactivation and cytotoxicity of the antitumor agent 2,5-diaziridinyl-3-(hydroxymethyl)-6-methyl-1,4-benzoquinone (RH1).
|
| |
Mol Pharmacol,
74,
1657-1665.
|
 |
|
|
|
|
 |
J.A.Boutin,
E.Marcheteau,
P.Hennig,
N.Moulharat,
S.Berger,
P.Delagrange,
J.P.Bouchet,
and
G.Ferry
(2008).
MT3/QR2 melatonin binding site does not use melatonin as a substrate or a co-substrate.
|
| |
J Pineal Res,
45,
524-531.
|
 |
|
|
|
|
 |
J.Brouillette,
and
R.Quirion
(2008).
Transthyretin: a key gene involved in the maintenance of memory capacities during aging.
|
| |
Neurobiol Aging,
29,
1721-1732.
|
 |
|
|
|
|
 |
L.Pirola,
and
S.Fröjdö
(2008).
Resveratrol: one molecule, many targets.
|
| |
IUBMB Life,
60,
323-332.
|
 |
|
|
|
|
 |
P.L.Toogood
(2008).
Mitochondrial drugs.
|
| |
Curr Opin Chem Biol,
12,
457-463.
|
 |
|
|
|
|
 |
R.Jockers,
P.Maurice,
J.A.Boutin,
and
P.Delagrange
(2008).
Melatonin receptors, heterodimerization, signal transduction and binding sites: what's new?
|
| |
Br J Pharmacol,
154,
1182-1195.
|
 |
|
|
|
|
 |
T.C.Hsieh,
Z.Wang,
H.Deng,
and
J.M.Wu
(2008).
Identification of glutathione sulfotransferase-pi (GSTP1) as a new resveratrol targeting protein (RTP) and studies of resveratrol-responsive protein changes by resveratrol affinity chromatography.
|
| |
Anticancer Res,
28,
29-36.
|
 |
|
|
|
|
 |
Y.Fu,
L.Buryanovskyy,
and
Z.Zhang
(2008).
Quinone reductase 2 is a catechol quinone reductase.
|
| |
J Biol Chem,
283,
23829-23835.
|
 |
|
|
|
|
 |
J.R.Gledhill,
M.G.Montgomery,
A.G.Leslie,
and
J.E.Walker
(2007).
Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols.
|
| |
Proc Natl Acad Sci U S A,
104,
13632-13637.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
U.Rix,
O.Hantschel,
G.Dürnberger,
L.L.Remsing Rix,
M.Planyavsky,
N.V.Fernbach,
I.Kaupe,
K.L.Bennett,
P.Valent,
J.Colinge,
T.Köcher,
and
G.Superti-Furga
(2007).
Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets.
|
| |
Blood,
110,
4055-4063.
|
 |
|
|
|
|
 |
Y.Hu,
S.Rahlfs,
V.Mersch-Sundermann,
and
K.Becker
(2007).
Resveratrol modulates mRNA transcripts of genes related to redox metabolism and cell proliferation in non-small-cell lung carcinoma cells.
|
| |
Biol Chem,
388,
207-219.
|
 |
|
|
|
|
 |
J.A.Baur,
and
D.A.Sinclair
(2006).
Therapeutic potential of resveratrol: the in vivo evidence.
|
| |
Nat Rev Drug Discov,
5,
493-506.
|
 |
|
|
|
|
 |
A.Slominski,
T.W.Fischer,
M.A.Zmijewski,
J.Wortsman,
I.Semak,
B.Zbytek,
R.M.Slominski,
and
D.J.Tobin
(2005).
On the role of melatonin in skin physiology and pathology.
|
| |
Endocrine,
27,
137-148.
|
 |
|
|
|
|
 |
J.A.Boutin,
F.Chatelain-Egger,
F.Vella,
P.Delagrange,
and
G.Ferry
(2005).
Quinone reductase 2 substrate specificity and inhibition pharmacology.
|
| |
Chem Biol Interact,
151,
213-228.
|
 |
|
|
|
|
 |
J.A.Boutin,
V.Audinot,
G.Ferry,
and
P.Delagrange
(2005).
Molecular tools to study melatonin pathways and actions.
|
| |
Trends Pharmacol Sci,
26,
412-419.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

