 |
PDBsum entry 1r6d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.46
- dTDP-glucose 4,6-dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
6-Deoxyhexose Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dTDP-alpha-D-glucose = dTDP-4-dehydro-6-deoxy-alpha-D-glucose + H2O
|
 |
 |
 |
 |
 |
dTDP-alpha-D-glucose
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
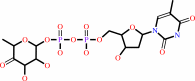 dTDP-4-dehydro-6-deoxy-alpha-D-glucose
dTDP-4-dehydro-6-deoxy-alpha-D-glucose
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
NAD(+)
|
 |
 |
 |
 |
 |
NAD(+)
Bound ligand (Het Group name =
NAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:2211-2220
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
High resolution X-ray structure of dTDP-glucose 4,6-dehydratase from Streptomyces venezuelae.
|
|
S.T.Allard,
W.W.Cleland,
H.M.Holden.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Desosamine is a 3-(dimethylamino)-3,4,6-trideoxyhexose found in some macrolide
antibiotics. In Streptomyces venezuelae, there are seven genes required for the
biosynthesis of this unusual sugar. One of the genes, desIV, codes for a
dTDP-glucose 4,6-dehydratase, which is referred to as DesIV. The reaction
mechanisms for these types of dehydratases are quite complicated with proton
abstraction from the sugar 4'-hydroxyl group and hydride transfer to NAD+,
proton abstraction at C-5, and elimination of the hydroxyl group at C-6 of the
sugar, and finally return of a proton to C-5 and a hydride from NADH to C-6.
Here we describe the cloning, overexpression, and purification, and high
resolution x-ray crystallographic analysis to 1.44 A of wild-type DesIV
complexed with dTDP. Additionally, for this study, a double site-directed mutant
protein (D128N/E129Q) was prepared, crystallized as a complex with NAD+ and the
substrate dTDP-glucose and its structure determined to 1.35 A resolution. In
DesIV, the phenolate group of Tyr(151) and O(gamma) of Thr(127) lie at 2.7 and
2.6 A, respectively from the 4'-hydroxyl group of the dTDP-glucose substrate.
The side chain of Asp(128) is in the correct position to function as a general
acid for proton donation to the 6'-hydroxyl group while the side chain of
Glu(129) is ideally situated to serve as the general base for proton abstraction
at C-5. This investigation provides further detailed information for
understanding the exquisite chemistry that occurs in these remarkable enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIG. 2. Ribbon representation of DesIV. Shown in a is a
ribbon representation of one subunit of DesIV with the bound
ligands, NAD^+ and dTDP, shown in a ball-and-stick
representation. The dimeric form of the enzyme is presented in b.
|
 |
Figure 3.
FIG. 3. Close up view of the DesIV active site. A close up
view of the active site, within  3.2 Å of the NAD^+
and dTDP ligands, is shown in a. For the sake of clarity, Ser87
and His88 were omitted from the figure. The side chain hydroxyl
group of Ser87 interacts with a phosphoryl oxygen of the NAD^+
while N 3.2 Å of the NAD^+
and dTDP ligands, is shown in a. For the sake of clarity, Ser87
and His88 were omitted from the figure. The side chain hydroxyl
group of Ser87 interacts with a phosphoryl oxygen of the NAD^+
while N  2 of His88 forms a
hydrogen bond with a phosphoryl oxygen of the dTDP. A schematic
of the hydrogen-bonding pattern around dTDP is displayed in b.
Possible hydrogen-bonding interactions within 2 of His88 forms a
hydrogen bond with a phosphoryl oxygen of the dTDP. A schematic
of the hydrogen-bonding pattern around dTDP is displayed in b.
Possible hydrogen-bonding interactions within  3.2
Å are indicated by dashed lines. 3.2
Å are indicated by dashed lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
2211-2220)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Pinta,
K.A.Duda,
A.Hanuszkiewicz,
Z.Kaczyński,
B.Lindner,
W.L.Miller,
H.Hyytiäinen,
C.Vogel,
S.Borowski,
K.Kasperkiewicz,
J.S.Lam,
J.Radziejewska-Lebrecht,
M.Skurnik,
and
O.Holst
(2009).
Identification and role of a 6-deoxy-4-keto-hexosamine in the lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3.
|
| |
Chemistry,
15,
9747-9754.
|
 |
|
|
|
|
 |
Q.Zhang,
F.Gao,
H.Peng,
H.Cheng,
Y.Liu,
J.Tang,
J.Thompson,
G.Wei,
J.Zhang,
Y.Du,
J.Yan,
and
G.F.Gao
(2009).
Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism.
|
| |
J Bacteriol,
191,
5832-5837.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.L.Chen,
Y.H.Chen,
Y.C.Lin,
K.C.Tsai,
and
H.T.Chiu
(2009).
Functional characterization and substrate specificity of spinosyn rhamnosyltransferase by in vitro reconstitution of spinosyn biosynthetic enzymes.
|
| |
J Biol Chem,
284,
7352-7363.
|
 |
|
|
|
|
 |
C.J.Thibodeaux,
C.E.Melançon,
and
H.W.Liu
(2008).
Natural-product sugar biosynthesis and enzymatic glycodiversification.
|
| |
Angew Chem Int Ed Engl,
47,
9814-9859.
|
 |
|
|
|
|
 |
E.S.Burgie,
and
H.M.Holden
(2007).
Molecular architecture of DesI: a key enzyme in the biosynthesis of desosamine.
|
| |
Biochemistry,
46,
8999-9006.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.D.King,
N.J.Harmer,
A.Preston,
C.M.Palmer,
M.Rejzek,
R.A.Field,
T.L.Blundell,
and
D.J.Maskell
(2007).
Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
|
| |
J Mol Biol,
374,
749-763.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.N.Hung,
E.Rangarajan,
C.Munger,
G.Nadeau,
T.Sulea,
and
A.Matte
(2006).
Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis.
|
| |
J Bacteriol,
188,
5606-5617.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.M.Koropatkin,
and
H.M.Holden
(2005).
Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi complexed with CDP-D-xylose.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
365-373.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

