 |
PDBsum entry 1pl3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1pl3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Cytochrome domain of cellobiose dehydrogenase, m65h mutant
|
|
Structure:
|
 |
Cellobiose dehydrogenase. Chain: a, b. Fragment: cytochrome type b heme domain. Synonym: cdh, cellobiose-quinone oxidoreductase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Phanerochaete chrysosporium. Organism_taxid: 5306. Gene: cdh-1 and cdh-2. Expressed in: phanerochaete chrysosporium. Expression_system_taxid: 5306.
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.174
|
R-free:
|
0.198
|
|
|
Authors:
|
 |
F.A.J.Rotsaert,B.M.Hallberg,S.De Vries,P.Moenne-Loccoz,C.Divne, M.H.Gold
|
Key ref:

|
 |
F.A.Rotsaert
et al.
(2003).
Biophysical and structural analysis of a novel heme B iron ligation in the flavocytochrome cellobiose dehydrogenase.
J Biol Chem,
278,
33224-33231.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
06-Jun-03
|
Release date:
|
01-Jul-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q01738
(CDH_PHACH) -
Cellobiose dehydrogenase from Phanerodontia chrysosporium
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
773 a.a.
186 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.99.18
- cellobiose dehydrogenase (acceptor).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-cellobiose + A = D-cellobiono-1,5-lactone + AH2
|
 |
 |
 |
 |
 |
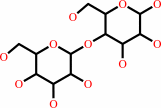 Cellobiose
Cellobiose
|
+
|
 acceptor
acceptor
|
=
|
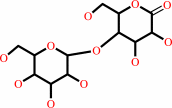 cellobiono-1,5-lactone
cellobiono-1,5-lactone
|
+
|
reduced acceptor
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Heme
|
 |
 |
 |
 |
 |
 FAD
FAD
|
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:33224-33231
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Biophysical and structural analysis of a novel heme B iron ligation in the flavocytochrome cellobiose dehydrogenase.
|
|
F.A.Rotsaert,
B.M.Hallberg,
S.de Vries,
P.Moenne-Loccoz,
C.Divne,
V.Renganathan,
M.H.Gold.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The fungal extracellular flavocytochrome cellobiose dehydrogenase (CDH)
participates in lignocellulose degradation. The enzyme has a cytochrome domain
connected to a flavin-binding domain by a peptide linker. The cytochrome domain
contains a 6-coordinate low spin b-type heme with unusual iron ligands and
coordination geometry. Wild type CDH is only the second example of a b-type heme
with Met-His ligation, and it is the first example of a Met-His ligation of heme
b where the ligands are arranged in a nearly perpendicular orientation. To
investigate the ligation further, Met65 was replaced with a histidine to create
a bis-histidyl ligated iron typical of b-type cytochromes. The variant is
expressed as a stable 90-kDa protein that retains the flavin domain catalytic
reactivity. However, the ability of the mutant to reduce external one-electron
acceptors such as cytochrome c is impaired. Electrochemical measurements
demonstrate a decrease in the redox midpoint potential of the heme by 210 mV. In
contrast to the wild type enzyme, the ferric state of the protoheme displays a
mixed low spin/high spin state at room temperature and low spin character at 90
K, as determined by resonance Raman spectroscopy. The wild type cytochrome does
not bind CO, but the ferrous state of the variant forms a CO complex, although
the association rate is very low. The crystal structure of the M65H cytochrome
domain has been determined at 1.9 A resolution. The variant structure confirms a
bis-histidyl ligation but reveals unusual features. As for the wild type enzyme,
the ligands have a nearly perpendicular arrangement. Furthermore, the iron is
bound by imidazole N delta 1 and N epsilon 2 nitrogen atoms, rather than the
typical N epsilon 2/N epsilon 2 coordination encountered in bis-histidyl ligated
heme proteins. To our knowledge, this is the first example of a bis-histidyl N
delta 1/N epsilon 2-coordinated protoporphyrin IX iron.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIG. 3. High frequency RR spectra of the oxidized rCDH and
the M65H variant, obtained at room temperature (RT, A) and 90
K(B), with Soret excitation (413 nm) in 20 mM sodium succinate,
pH 4.5.
|
 |
Figure 5.
FIG. 5. a, the  [A]-weighted F[o] -
F[c] electron density around the iron-protoporphyrin IX ring in
the M65H cytochrome domain. The 6-coordinate heme iron is
ligated by His65 N [A]-weighted F[o] -
F[c] electron density around the iron-protoporphyrin IX ring in
the M65H cytochrome domain. The 6-coordinate heme iron is
ligated by His65 N  1 and His163 N 1 and His163 N  2. b,
superposition of the M65H variant (gold) and wild type (green)
heme-binding pocket (Protein Data Bank code 1D7C [PDB]
) (7). The drawings were made with the program Pymol
(pymol.sourceforge.net). 2. b,
superposition of the M65H variant (gold) and wild type (green)
heme-binding pocket (Protein Data Bank code 1D7C [PDB]
) (7). The drawings were made with the program Pymol
(pymol.sourceforge.net).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
33224-33231)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

