 |
PDBsum entry 1p3h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Mycobacterium tuberculosis chaperonin 10 heptamers self-Associate through their biologically active loops.
|
 |
|
Authors
|
 |
M.M.Roberts,
A.R.Coker,
G.Fossati,
P.Mascagni,
A.R.Coates,
S.P.Wood.
|
 |
|
Ref.
|
 |
J Bacteriol, 2003,
185,
4172-4185.
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
The crystal structure of Mycobacterium tuberculosis chaperonin 10 (cpn10(Mt))
has been determined to a resolution of 2.8 A. Two dome-shaped cpn10(Mt)
heptamers complex through loops at their bases to form a tetradecamer with 72
symmetry and a spherical cage-like structure. The hollow interior enclosed by
the tetradecamer is lined with hydrophilic residues and has dimensions of 30 A
perpendicular to and 60 A along the sevenfold axis. Tetradecameric cpn10(Mt) has
also been observed in solution by dynamic light scattering. Through its base
loop sequence cpn10(Mt) is known to be the agent in the bacterium responsible
for bone resorption and for the contribution towards its strong T-cell
immunogenicity. Superimposition of the cpn10(Mt) sequences 26 to 32 and 66 to 72
and E. coli GroES 25 to 31 associated with bone resorption activity shows them
to have similar conformations and structural features, suggesting that there may
be a common receptor for the bone resorption sequences. The base loops of cpn10s
in general also attach to the corresponding chaperonin 60 (cpn60) to enclose
unfolded protein and to facilitate its correct folding in vivo. Electron density
corresponding to a partially disordered protein subunit appears encapsulated
within the interior dome cavity of each heptamer. This suggests that the binding
of substrates to cpn10 is possible in the absence of cpn60.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Crystallization, X-Ray diffraction and preliminary structure analysis of mycobacterium tuberculosis chaperonin 10.
|
 |
|
Authors
|
 |
M.M.Roberts,
A.R.Coker,
G.Fossati,
P.Mascagni,
A.R.Coates,
S.P.Wood.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 1999,
55,
910-914.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 An X-ray diffraction image taken at CCLRC Daresbury
Laboratory on station 7.2, recorded on a large MAR Research
image plate with an X-ray wavelength of 1.488 and
crystal-to-detector distance of 230 mm. The circles indicate
diffraction limits of 11.2, 5.6, 3.7 and 2.8 Å, and a
rotation angle of 1° was used.
|
 |
Figure 2.
Figure 2 Stereographic projections of self-rotation functions
produced by POLARRFN with normalized data (E values) from 10 to
5.1 Å resolution with a 30 Å integration radius.
(a) and (c) illustrate the twofold axes on  =
180°. The peaks corresponding to the crystallographic 2[1]
axis can be seen at the perimeter of the plot ( =
180°. The peaks corresponding to the crystallographic 2[1]
axis can be seen at the perimeter of the plot (  =
90°) and occur at =
90°) and occur at 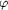 values
of 90 and -90°. Two strings of seven unique NCS twofold axes
are generated from each double heptamer in the unit cell. These
appear as arcs above and below the equator and reflect the tilt
in the plane of each double ring with respect to the xz plane
owing to their positioning about the 2[1] axis. The NCS twofold
axes from each heptamer coincide to give a prominent NCS axis on
the right-hand side. Together with the crystallographic 2[1]
axis, this generates an orthogonal NCS axis as a peak close to
the centre of the projection ( values
of 90 and -90°. Two strings of seven unique NCS twofold axes
are generated from each double heptamer in the unit cell. These
appear as arcs above and below the equator and reflect the tilt
in the plane of each double ring with respect to the xz plane
owing to their positioning about the 2[1] axis. The NCS twofold
axes from each heptamer coincide to give a prominent NCS axis on
the right-hand side. Together with the crystallographic 2[1]
axis, this generates an orthogonal NCS axis as a peak close to
the centre of the projection (  =
0°). (b) and (d) illustrate the sevenfold axes on =
0°). (b) and (d) illustrate the sevenfold axes on  =
51°. The displacement of each peak from the perimeter at =
51°. The displacement of each peak from the perimeter at
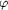 =
90 and -90° represents the 35° tilt of the NCS sevenfold
axis of each double heptamer related by 2[1] symmetry from the y
axis. (a) and (b) represent the experimentally collected data,
whilst (c) and (d) represent data back-transformed from the
initial molecular-replacement search model rigid-body refined to
2.8 Å. =
90 and -90° represents the 35° tilt of the NCS sevenfold
axis of each double heptamer related by 2[1] symmetry from the y
axis. (a) and (b) represent the experimentally collected data,
whilst (c) and (d) represent data back-transformed from the
initial molecular-replacement search model rigid-body refined to
2.8 Å.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the IUCr
|
 |
|
Secondary reference #2
|
 |
|
Title
|
 |
Mycobacterium tuberculosis chaperonin 10 stimulates bone resorption: a potential contributory factor in pott'S disease.
|
 |
|
Authors
|
 |
S.Meghji,
P.A.White,
S.P.Nair,
K.Reddi,
K.Heron,
B.Henderson,
A.Zaliani,
G.Fossati,
P.Mascagni,
J.F.Hunt,
M.M.Roberts,
A.R.Coates.
|
 |
|
Ref.
|
 |
J Exp Med, 1997,
186,
1241-1246.
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
|

|
|
|
 |

