 |
PDBsum entry 1n4f

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.17
- lysozyme.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Hydrolysis of the 1,4-beta-linkages between N-acetyl-D-glucosamine and N-acetylmuramic acid in peptidoglycan heteropolymers of the prokaryotes cell walls.
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
59:887-896
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Phasing power at the K absorption edge of organic arsenic.
|
|
P.Retailleau,
T.Prangé.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Single/multiple-wavelength anomalous dispersion (SAD/MAD) experiments were
performed on a crystal of an organic arsenic derivative of hen egg-white
lysozyme. A para-arsanilate compound used as a crystallizing reagent was
incorporated into the ordered solvent region of the lysozyme molecule.
Diffraction data were collected to high resolution (</=2.0 A) at three
wavelengths around the K edge (1.04 A) of arsenic at beamline BM30A, ESRF
synchrotron. Anomalous Patterson maps clearly showed the main arsanilate site to
be between three symmetry-related lysozyme molecules, at a location previously
occupied by a para-toluenesulfonate anion. MAD phases at 2 A derived using the
program SHARP led to an electron-density map of sufficient quality to start
manual building of the protein model. Amplitudes from a second crystal measured
to a resolution of 1.8 A at the peak wavelength revealed two additional
heavy-atom sites, which reinforced the anomalous subset model and therefore
dramatically improved the phasing power of the arsenic derivative. The
subsequent solvent-flattened map was of such high accuracy that the program
ARP/wARP was able to build a nearly complete model automatically. This work
emphasizes the great potential of arsenic for de novo structure determination
using anomalous dispersion methods.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2 Resolution-dependent statistics of the anomalous signal
and of quality indicators for two different data sets (blue for
crystal 1 and black for crystal 2): | 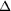 F[anom]|/ F[anom]|/
 ( (
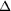 F[anom])
and I/ F[anom])
and I/  (I)
on the left y axis are absolute numbers, whereas R[sym] and | (I)
on the left y axis are absolute numbers, whereas R[sym] and |
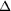 F[anom]|/F
are measured in percentages on the right y axis. F[anom]|/F
are measured in percentages on the right y axis.
|
 |
Figure 3.
Figure 3 {w = 1/2, 0 [216]<= u [217]<= 1/2, 0 [218]<= v [219]<=
1/2} Harker sections of anomalous difference Patterson maps for
(a) crystal 1 and (b) crystal 2. Levels are contoured in 1
[220][sigma] steps starting at 2 [221][sigma] .
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2003,
59,
887-896)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

