 |
PDBsum entry 1n07

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.26
- riboflavin kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
riboflavin + ATP = FMN + ADP + H+
|
 |
 |
 |
 |
 |
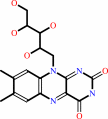 riboflavin
riboflavin
|
+
|
 ATP
ATP
|
=
|
FMN
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
ADP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+) or Zn(2+) or Mn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
326:1463-1473
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Schizosaccharomyces pombe riboflavin kinase reveals a novel ATP and riboflavin-binding fold.
|
|
S.Bauer,
K.Kemter,
A.Bacher,
R.Huber,
M.Fischer,
S.Steinbacher.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The essential redox cofactors riboflavin monophosphate (FMN) and flavin adenine
dinucleotide (FAD) are synthesised from their precursor, riboflavin, in
sequential reactions by the metal-dependent riboflavin kinase and FAD
synthetase. Here, we describe the 1.6A crystal structure of the
Schizosaccharomyces pombe riboflavin kinase. The enzyme represents a novel
family of phosphoryl transferring enzymes. It is a monomer comprising a central
beta-barrel clasped on one side by two C-terminal helices that display an L-like
shape. The opposite side of the beta-barrel serves as a platform for substrate
binding as demonstrated by complexes with ADP and FMN. Formation of the
ATP-binding site requires significant rearrangements in a short alpha-helix as
compared to the substrate free form. The diphosphate moiety of ADP is covered by
the glycine-rich flap I formed from parts of this alpha-helix. In contrast, no
significant changes are observed upon binding of riboflavin. The ribityl
side-chain might be covered by a rather flexible flap II. The unusual
metal-binding site involves, in addition to the ADP phosphates, only the
strictly conserved Thr45. This may explain the preference for zinc observed in
vitro.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Reaction catalysed by riboflavin kinase and FAD
synthetase. 1, Riboflavin; 2, FMN; 3, FAD.
|
 |
Figure 5.
Figure 5. Proposed catalytic mechanism of riboflavin
synthase. (a) Stereo drawings of the active site of the
riboflavin kinase. Bound FMN and ADP are shown as ball-and-stick
models. The position of the zinc ion was taken from ADP+Zn
complex. Flap I covers the b and g phosphate-binding sites. (b)
Residues involved in catalysis.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2003,
326,
1463-1473)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Yruela,
S.Arilla-Luna,
M.Medina,
and
B.Contreras-Moreira
(2010).
Evolutionary divergence of chloroplast FAD synthetase proteins.
|
| |
BMC Evol Biol,
10,
311.
|
 |
|
|
|
|
 |
S.Frago,
A.Velázquez-Campoy,
and
M.Medina
(2009).
The Puzzle of Ligand Binding to Corynebacterium ammoniagenes FAD Synthetase.
|
| |
J Biol Chem,
284,
6610-6619.
|
 |
|
|
|
|
 |
T.A.Giancaspero,
V.Locato,
M.C.de Pinto,
L.De Gara,
and
M.Barile
(2009).
The occurrence of riboflavin kinase and FAD synthetase ensures FAD synthesis in tobacco mitochondria and maintenance of cellular redox status.
|
| |
FEBS J,
276,
219-231.
|
 |
|
|
|
|
 |
F.J.Sandoval,
Y.Zhang,
and
S.Roje
(2008).
Flavin Nucleotide Metabolism in Plants: MONOFUNCTIONAL ENZYMES SYNTHESIZE FAD IN PLASTIDS.
|
| |
J Biol Chem,
283,
30890-30900.
|
 |
|
|
|
|
 |
S.Frago,
M.Martínez-Júlvez,
A.Serrano,
and
M.Medina
(2008).
Structural analysis of FAD synthetase from Corynebacterium ammoniagenes.
|
| |
BMC Microbiol,
8,
160.
|
 |
|
|
|
|
 |
Z.Mashhadi,
H.Zhang,
H.Xu,
and
R.H.White
(2008).
Identification and characterization of an archaeon-specific riboflavin kinase.
|
| |
J Bacteriol,
190,
2615-2618.
|
 |
|
|
|
|
 |
A.K.Hirsch,
F.R.Fischer,
and
F.Diederich
(2007).
Phosphate recognition in structural biology.
|
| |
Angew Chem Int Ed Engl,
46,
338-352.
|
 |
|
|
|
|
 |
M.Ammelburg,
M.D.Hartmann,
S.Djuranovic,
V.Alva,
K.K.Koretke,
J.Martin,
G.Sauer,
V.Truffault,
K.Zeth,
A.N.Lupas,
and
M.Coles
(2007).
A CTP-dependent archaeal riboflavin kinase forms a bridge in the evolution of cradle-loop barrels.
|
| |
Structure,
15,
1577-1590.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Eisenreich,
M.Joshi,
B.Illarionov,
G.Richter,
W.Römisch-Margl,
F.Müller,
A.Bacher,
and
M.Fischer
(2007).
13C Isotopologue editing of FMN bound to phototropin domains.
|
| |
FEBS J,
274,
5876-5890.
|
 |
|
|
|
|
 |
F.J.Sandoval,
and
S.Roje
(2005).
An FMN hydrolase is fused to a riboflavin kinase homolog in plants.
|
| |
J Biol Chem,
280,
38337-38345.
|
 |
|
|
|
|
 |
M.Fischer,
and
A.Bacher
(2005).
Biosynthesis of flavocoenzymes.
|
| |
Nat Prod Rep,
22,
324-350.
|
 |
|
|
|
|
 |
S.Cheek,
K.Ginalski,
H.Zhang,
and
N.V.Grishin
(2005).
A comprehensive update of the sequence and structure classification of kinases.
|
| |
BMC Struct Biol,
5,
6.
|
 |
|
|
|
|
 |
W.Wang,
R.Kim,
H.Yokota,
and
S.H.Kim
(2005).
Crystal structure of flavin binding to FAD synthetase of Thermotoga maritima.
|
| |
Proteins,
58,
246-248.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Cheek,
Y.Qi,
S.S.Krishna,
L.N.Kinch,
and
N.V.Grishin
(2004).
4SCOPmap: automated assignment of protein structures to evolutionary superfamilies.
|
| |
BMC Bioinformatics,
5,
197.
|
 |
|
|
|
|
 |
S.Karthikeyan,
Q.Zhou,
A.L.Osterman,
and
H.Zhang
(2003).
Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism.
|
| |
Biochemistry,
42,
12532-12538.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Steinbacher,
S.Schiffmann,
G.Richter,
R.Huber,
A.Bacher,
and
M.Fischer
(2003).
Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with divalent metal ions and the substrate ribulose 5-phosphate: implications for the catalytic mechanism.
|
| |
J Biol Chem,
278,
42256-42265.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

