 |
PDBsum entry 1kl7

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of threonine synthase from yeast
|
|
Structure:
|
 |
Threonine synthase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.201
|
R-free:
|
0.253
|
|
|
Authors:
|
 |
M.Garrido-Franco,S.Ehlert,A.Messerschmidt,S.Marinkovic,R.Huber, B.Laber,G.P.Bourenkov,T.Clausen
|
Key ref:

|
 |
M.Garrido-Franco
et al.
(2002).
Structure and function of threonine synthase from yeast.
J Biol Chem,
277,
12396-12405.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
11-Dec-01
|
Release date:
|
24-Apr-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P16120
(THRC_YEAST) -
Threonine synthase from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
514 a.a.
509 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.3.1
- threonine synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Threonine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-phospho-L-homoserine + H2O = L-threonine + phosphate
|
 |
 |
 |
 |
 |
 O-phospho-L-homoserine
O-phospho-L-homoserine
|
+
|
H2O
|
=
|
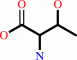 L-threonine
L-threonine
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:12396-12405
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and function of threonine synthase from yeast.
|
|
M.Garrido-Franco,
S.Ehlert,
A.Messerschmidt,
S.Marinkovic',
R.Huber,
B.Laber,
G.P.Bourenkov,
T.Clausen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Threonine synthase catalyzes the final step of threonine biosynthesis, the
pyridoxal 5'-phosphate (PLP)-dependent conversion of O-phosphohomoserine into
threonine and inorganic phosphate. Threonine is an essential nutrient for
mammals, and its biosynthetic machinery is restricted to bacteria, plants, and
fungi; therefore, threonine synthase represents an interesting pharmaceutical
target. The crystal structure of threonine synthase from Saccharomyces
cerevisiae has been solved at 2.7 A resolution using multiwavelength anomalous
diffraction. The structure reveals a monomer as active unit, which is subdivided
into three distinct domains: a small N-terminal domain, a PLP-binding domain
that covalently anchors the cofactor and a so-called large domain, which
contains the main of the protein body. All three domains show the typical open
alpha/beta architecture. The cofactor is bound at the interface of all three
domains, buried deeply within a wide canyon that penetrates the whole molecule.
Based on structural alignments with related enzymes, an enzyme-substrate complex
was modeled into the active site of yeast threonine synthase, which revealed
essentials for substrate binding and catalysis. Furthermore, the comparison with
related enzymes of the beta-family of PLP-dependent enzymes indicated structural
determinants of the oligomeric state and thus rationalized for the first time
how a PLP enzyme acts in monomeric form.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Active site of yTS. a, schematic representation
of the functionally important interactions within the active
site. On the left side, hydrogen bonds between protein, water
(dark balls) and cofactor (light gray) are indicated, whereas
the fixation of the PLP pyridine ring is shown on the right
side. b, detailed active site architecture. The internal
aldimine is seen in light gray, water molecules are shown as
dark balls, and the macrodipole of helix  10 is
indicated. b was produced with DINO. 10 is
indicated. b was produced with DINO.
|
 |
Figure 5.
Fig. 5. Mechanistic features of yTS. a, drawing of the
modeled external aldimine between OPHS and PLP (light gray).
Hydrogen bonds and interatomic distances (Å) relevant for
substrate binding are indicated. b, mechanism of the reaction
catalyzed by yTS where the substrate OPHS is converted to
inorganic phosphate and threonine. The precise electron
movements are indicated with arrows.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
12396-12405)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.E.Graham,
S.M.Taylor,
R.Z.Wolf,
and
S.C.Namboori
(2009).
Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase.
|
| |
Biochem J,
424,
467-478.
|
 |
|
|
|
|
 |
G.K.Smith,
Z.Ke,
A.C.Hengge,
D.Xu,
D.Xie,
and
H.Guo
(2009).
Active-site dynamics of SpvC virulence factor from Salmonella typhimurium and density functional theory study of phosphothreonine lyase catalysis.
|
| |
J Phys Chem B,
113,
15327-15333.
|
 |
|
|
|
|
 |
M.Goto,
T.Yamauchi,
N.Kamiya,
I.Miyahara,
T.Yoshimura,
H.Mihara,
T.Kurihara,
K.Hirotsu,
and
N.Esaki
(2009).
Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe.
|
| |
J Biol Chem,
284,
25944-25952.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zhu,
H.Li,
C.Long,
L.Hu,
H.Xu,
L.Liu,
S.Chen,
D.C.Wang,
and
F.Shao
(2007).
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase.
|
| |
Mol Cell,
28,
899-913.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.A.Azevedo,
M.Lancien,
and
P.J.Lea
(2006).
The aspartic acid metabolic pathway, an exciting and essential pathway in plants.
|
| |
Amino Acids,
30,
143-162.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

