 |
PDBsum entry 1kb0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1kb0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.9.1
- alcohol dehydrogenase (azurin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 oxidized [azurin] + a primary alcohol = 2 reduced [azurin] + an aldehyde + 2 H+
|
 |
 |
 |
 |
 |
2
×
oxidized [azurin]
|
+
|
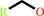 primary alcohol
primary alcohol
|
=
|
2
×
reduced [azurin]
|
+
|
 aldehyde
aldehyde
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme; Pyrroloquinoline quinone
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEC)
matches with 95.45% similarity
|
Pyrroloquinoline quinone
Bound ligand (Het Group name =
PQQ)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:3727-3732
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of quinohemoprotein alcohol dehydrogenase from Comamonas testosteroni: structural basis for substrate oxidation and electron transfer.
|
|
A.Oubrie,
H.J.Rozeboom,
K.H.Kalk,
E.G.Huizinga,
B.W.Dijkstra.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Quinoprotein alcohol dehydrogenases are redox enzymes that participate in
distinctive catabolic pathways that enable bacteria to grow on various alcohols
as the sole source of carbon and energy. The x-ray structure of the
quinohemoprotein alcohol dehydrogenase from Comamonas testosteroni has been
determined at 1.44 A resolution. It comprises two domains. The N-terminal domain
has a beta-propeller fold and binds one pyrroloquinoline quinone cofactor and
one calcium ion in the active site. A tetrahydrofuran-2-carboxylic acid molecule
is present in the substrate-binding cleft. The position of this oxidation
product provides valuable information on the amino acid residues involved in the
reaction mechanism and their function. The C-terminal domain is an alpha-helical
type I cytochrome c with His(608) and Met(647) as heme-iron ligands. This is the
first reported structure of an electron transfer system between a quinoprotein
alcohol dehydrogenase and cytochrome c. The shortest distance between
pyrroloquinoline quinone and heme c is 12.9 A, one of the longest physiological
edge-to-edge distances yet determined between two redox centers. A highly
unusual disulfide bond between two adjacent cysteines bridges the redox centers.
It appears essential for electron transfer. A water channel delineates a
possible pathway for proton transfer from the active site to the solvent.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. Proposed reaction mechanisms for substrate
conversion by QH-ADH. A, alcohol oxidation. The reactive C5 atom
of PQQ is indicated by the number 5. B, aldehyde oxidation.
|
 |
Figure 5.
Fig. 5. Proposed pathways for electron and proton
transfer. A, electron transfer. Optimal pathways and one longer
alternative are indicated. B, proton transfer. Hydrogen bonds
are visualized by black dotted lines. The gap in the
hydrogen-bonding pattern is indicated. Residues possibly
involved in either pathway are shown as ball-and-stick models.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
3727-3732)
copyright 2002.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Li,
J.H.Gan,
F.S.Mathews,
and
Z.X.Xia
(2011).
The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase.
|
| |
Biochem Biophys Res Commun,
406,
621-626.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Mennenga,
C.W.Kay,
and
H.Görisch
(2009).
Quinoprotein ethanol dehydrogenase from Pseudomonas aeruginosa: the unusual disulfide ring formed by adjacent cysteine residues is essential for efficient electron transfer to cytochrome c550.
|
| |
Arch Microbiol,
191,
361-367.
|
 |
|
|
|
|
 |
M.J.Ellis,
J.G.Grossmann,
R.R.Eady,
and
S.S.Hasnain
(2007).
Genomic analysis reveals widespread occurrence of new classes of copper nitrite reductases.
|
| |
J Biol Inorg Chem,
12,
1119-1127.
|
 |
|
|
|
|
 |
Y.Wu,
B.Ilan,
and
G.A.Voth
(2007).
Charge delocalization in proton channels, II: the synthetic LS2 channel and proton selectivity.
|
| |
Biophys J,
92,
61-69.
|
 |
|
|
|
|
 |
P.A.Williams,
L.Coates,
F.Mohammed,
R.Gill,
P.T.Erskine,
A.Coker,
S.P.Wood,
C.Anthony,
and
J.B.Cooper
(2005).
The atomic resolution structure of methanol dehydrogenase from Methylobacterium extorquens.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
75-79.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Hudáky,
Z.Gáspári,
O.Carugo,
M.Cemazar,
S.Pongor,
and
A.Perczel
(2004).
Vicinal disulfide bridge conformers by experimental methods and by ab initio and DFT molecular computations.
|
| |
Proteins,
55,
152-168.
|
 |
|
|
|
|
 |
S.Y.Reddy,
and
T.C.Bruice
(2004).
Determination of enzyme mechanisms by molecular dynamics: studies on quinoproteins, methanol dehydrogenase, and soluble glucose dehydrogenase.
|
| |
Protein Sci,
13,
1965-1978.
|
 |
|
|
|
|
 |
H.Toyama,
T.Fujii,
N.Aoki,
K.Matsushita,
and
O.Adachi
(2003).
Molecular cloning of quinohemoprotein alcohol dehydrogenase, ADH IIB, from Pseudomonas putida HK5.
|
| |
Biosci Biotechnol Biochem,
67,
1397-1400.
|
 |
|
|
|
|
 |
Z.W.Chen,
K.Matsushita,
T.Yamashita,
T.A.Fujii,
H.Toyama,
O.Adachi,
H.D.Bellamy,
and
F.S.Mathews
(2002).
Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
|
| |
Structure,
10,
837-849.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

