 |
PDBsum entry 1js1

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of a new transcarbamylase from the anaerobic bacterium bacteroides fragilis at 2.0 a resolution
|
|
Structure:
|
 |
Transcarbamylase. Chain: x, y, z. Engineered: yes
|
|
Source:
|
 |
Bacteroides fragilis. Organism_taxid: 817. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Trimer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.206
|
R-free:
|
0.252
|
|
|
Authors:
|
 |
D.Shi,R.Gallegos,J.Deponte Iii,H.Morizono,X.Yu,N.M.Allewell,M.Malamy, M.Tuchman
|
Key ref:

|
 |
D.Shi
et al.
(2002).
Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution.
J Mol Biol,
320,
899-908.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
15-Aug-01
|
Release date:
|
17-Jul-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8A1E9
(AOTC_BACTN) -
N-succinylornithine carbamoyltransferase from Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
318 a.a.
324 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 22 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.11
- N-succinylornithine carbamoyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N2-succinyl-L-ornithine + carbamoyl phosphate = N2-succinyl-L- citrulline + phosphate + H+
|
 |
 |
 |
 |
 |
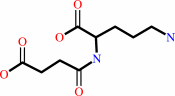 N(2)-succinyl-L-ornithine
N(2)-succinyl-L-ornithine
|
+
|
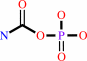 carbamoyl phosphate
carbamoyl phosphate
|
=
|
N(2)-succinyl-L- citrulline
|
+
|
 phosphate
phosphate
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
320:899-908
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution.
|
|
D.Shi,
R.Gallegos,
J.DePonte,
H.Morizono,
X.Yu,
N.M.Allewell,
M.Malamy,
M.Tuchman.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A transcarbamylase-like protein essential for arginine biosynthesis in the
anaerobic bacterium Bacteroides fragilis has been purified and crystallized in
space group P4(3)2(1)2 (a=b=153.4 A, c=94.8 A). The structure was solved using a
single isomorphous replacement with anomalous scattering (SIRAS) and was refined
at 2.0 A resolution to an R-factor of 20.6% (R-free=25.2%). The molecular model
is trimeric and comprises 960 amino acid residues, two phosphate groups and 422
water molecules. The monomer has the consensus transcarbamylase fold with two
structural domains linked by two long interdomain helices: the putative
carbamoyl phosphate-binding domain and a binding domain for the second
substrate. Each domain has a central parallel beta-sheet surrounded by
alpha-helices and loops with alpha/beta topology. The putative carbamoyl
phosphate-binding site is similar to those in ornithine transcarbamylases
(OTCases) and aspartate transcarbamylases (ATCases); however, the second
substrate-binding site is strikingly different. This site has several insertions
and deletions, and residues critical to substrate binding and catalysis in other
known transcarbamylases are not conserved. The three-dimensional structure and
the fact that this protein is essential for arginine biosynthesis suggest
strongly that it is a new member of the transcarbamylase family. A similar
protein has been found in Xylella fastidiosa, a bacterium that infects grapes,
citrus and other plants.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Ribbon diagram of the monomer. Green arrows
indicate segments that are hydrogen bonded to an adjacent strand
as required by the Kabsch & Sander[38] definition of b
character. a-Helices are light blue and 3[10] helices are dark
blue. The phosphate group is represented as a ball-and-stick
model. The Figure was drawn with RIBBONS.[39]
|
 |
Figure 4.
Figure 4. Comparison of the carbamoyl phosphate binding
site between the B. fragilis transcarbamylase-like structure and
human OTCase. (a) Putative carbamoyl phosphate-binding site in
the B. fragilis protein. The phosphate group and two active-site
water molecules are shown as a ball-and-stick model. (b)
Carbamoyl phosphate-binding site in human OTCase.[42] CP is
shown as a thick ball-and-stick model. The phosphate group in
the B. fragilis protein is located in the same position as the
phosphate moiety of CP in human OTCase and interacts with the
protein in a similar way. Two water molecules occupy the
respective positions of the carbamoyl nitrogen and oxygen atoms
of CP. The Figure was drawn with MOLSCRIPT.[41]
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
320,
899-908)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Bornschlögl,
D.M.Anstrom,
E.Mey,
J.Dzubiella,
M.Rief,
and
K.T.Forest
(2009).
Tightening the knot in phytochrome by single-molecule atomic force microscopy.
|
| |
Biophys J,
96,
1508-1514.
|
 |
|
|
|
|
 |
D.Shi,
X.Yu,
J.Cabrera-Luque,
T.Y.Chen,
L.Roth,
H.Morizono,
N.M.Allewell,
and
M.Tuchman
(2007).
A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
|
| |
Protein Sci,
16,
1689-1699.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Xu,
B.Labedan,
and
N.Glansdorff
(2007).
Surprising arginine biosynthesis: a reappraisal of the enzymology and evolution of the pathway in microorganisms.
|
| |
Microbiol Mol Biol Rev,
71,
36-47.
|
 |
|
|
|
|
 |
D.Shi,
H.Morizono,
J.Cabrera-Luque,
X.Yu,
L.Roth,
M.H.Malamy,
N.M.Allewell,
and
M.Tuchman
(2006).
Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
|
| |
J Biol Chem,
281,
20623-20631.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Shi,
X.Yu,
L.Roth,
H.Morizono,
M.Tuchman,
and
N.M.Allewell
(2006).
Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
|
| |
Proteins,
64,
532-542.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Morizono,
J.Cabrera-Luque,
D.Shi,
R.Gallegos,
S.Yamaguchi,
X.Yu,
N.M.Allewell,
M.H.Malamy,
and
M.Tuchman
(2006).
Acetylornithine transcarbamylase: a novel enzyme in arginine biosynthesis.
|
| |
J Bacteriol,
188,
2974-2982.
|
 |
|
|
|
|
 |
P.Virnau,
L.A.Mirny,
and
M.Kardar
(2006).
Intricate knots in proteins: Function and evolution.
|
| |
PLoS Comput Biol,
2,
e122.
|
 |
|
|
|
|
 |
Y.Xu,
N.Glansdorff,
and
B.Labedan
(2006).
Bioinformatic analysis of an unusual gene-enzyme relationship in the arginine biosynthetic pathway among marine gamma proteobacteria: implications concerning the formation of N-acetylated intermediates in prokaryotes.
|
| |
BMC Genomics,
7,
4.
|
 |
|
|
|
|
 |
D.Shi,
H.Morizono,
X.Yu,
L.Roth,
L.Caldovic,
N.M.Allewell,
M.H.Malamy,
and
M.Tuchman
(2005).
Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.
|
| |
J Biol Chem,
280,
14366-14369.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.G.Naumoff,
Y.Xu,
N.Glansdorff,
and
B.Labedan
(2004).
Retrieving sequences of enzymes experimentally characterized but erroneously annotated : the case of the putrescine carbamoyltransferase.
|
| |
BMC Genomics,
5,
52.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

