 |
PDBsum entry 1jr8

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1jr8
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.3.2
- thiol oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 R'C(R)SH + O2 = R'C(R)S-S(R)CR' + H2O2
|
 |
 |
 |
 |
 |
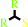 2
×
R'C(R)SH
2
×
R'C(R)SH
|
+
|
 O2
O2
|
=
|
 R'C(R)S-S(R)CR'
R'C(R)S-S(R)CR'
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
9:61-67
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p.
|
|
E.Gross,
C.S.Sevier,
A.Vala,
C.A.Kaiser,
D.Fass.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Erv2p is an FAD-dependent sulfhydryl oxidase that can promote disulfide bond
formation during protein biosynthesis in the yeast endoplasmic reticulum. The
structure of Erv2p, determined by X-ray crystallography to 1.5 A resolution,
reveals a helix-rich dimer with no global resemblance to other known FAD-binding
proteins or thiol oxidoreductases. Two pairs of cysteine residues are required
for Erv2p activity. The first (Cys-Gly-Glu-Cys) is adjacent to the isoalloxazine
ring of the FAD. The second (Cys-Gly-Cys) is part of a flexible C-terminal
segment that can swing into the vicinity of the first cysteine pair in the
opposite subunit of the dimer and may shuttle electrons between substrate
protein dithiols and the FAD-proximal disulfide.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. FAD binding site. a, A ribbon diagram of the
polypeptide backbone with some side chains and the bound FAD
shown in ball-and-stick representation. The bent conformation of
the FAD buries the isoalloxazine ring and adenine portions of
the cofactor while keeping the intervening regions surface
exposed. b, The ribbon trace of the polypeptide backbone has
been removed in this panel so that the residues involved in
aromatic ring stacking with the FAD can be identified.
|
 |
Figure 6.
Figure 6. Erv2p flexible C-terminal tail. The four molecules
in the asymmetric unit of the crystals grown from the original
Erv2-  N
preparation were superposed to compare the orientations of the
C-terminal regions containing the Cys-Gly-Cys sequence. In this
crystal, one of the four C-terminal tails packs against the
neighboring active site. The arrow indicates the significant
conformational differences between the superposed molecules.
Shown in the inset is a model of an intersubunit disulfide bond
constructed by changing the N
preparation were superposed to compare the orientations of the
C-terminal regions containing the Cys-Gly-Cys sequence. In this
crystal, one of the four C-terminal tails packs against the
neighboring active site. The arrow indicates the significant
conformational differences between the superposed molecules.
Shown in the inset is a model of an intersubunit disulfide bond
constructed by changing the  1
side chain torsion angles of Cys 178 and Cys 121 to decrease the
sulfur -sulfur bond distance between these residues to 2.03 Å. 1
side chain torsion angles of Cys 178 and Cys 121 to decrease the
sulfur -sulfur bond distance between these residues to 2.03 Å.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2002,
9,
61-67)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.R.Shouldice,
B.Heras,
P.M.Walden,
M.Totsika,
M.A.Schembri,
and
J.L.Martin
(2011).
Structure and function of DsbA, a key bacterial oxidative folding catalyst.
|
| |
Antioxid Redox Signal,
14,
1729-1760.
|
 |
|
|
|
|
 |
W.Zheng,
Y.Chu,
Q.Yin,
L.Xu,
C.Yang,
W.Zhang,
Y.Tang,
and
Y.Yang
(2011).
Crucial effect of the first CXXC motif of human QSOX 1b on the activity to different substrates.
|
| |
J Biochem,
149,
293-300.
|
 |
|
|
|
|
 |
D.P.Sideris,
and
K.Tokatlidis
(2010).
Oxidative protein folding in the mitochondrial intermembrane space.
|
| |
Antioxid Redox Signal,
13,
1189-1204.
|
 |
|
|
|
|
 |
E.Lionaki,
M.Aivaliotis,
C.Pozidis,
and
K.Tokatlidis
(2010).
The N-terminal shuttle domain of Erv1 determines the affinity for Mia40 and mediates electron transfer to the catalytic Erv1 core in yeast mitochondria.
|
| |
Antioxid Redox Signal,
13,
1327-1339.
|
 |
|
|
|
|
 |
E.Pedone,
D.Limauro,
K.D'Ambrosio,
G.De Simone,
and
S.Bartolucci
(2010).
Multiple catalytically active thioredoxin folds: a winning strategy for many functions.
|
| |
Cell Mol Life Sci,
67,
3797-3814.
|
 |
|
|
|
|
 |
J.M.Herrmann,
and
J.Riemer
(2010).
Oxidation and reduction of cysteines in the intermembrane space of mitochondria: multiple facets of redox control.
|
| |
Antioxid Redox Signal,
13,
1323-1326.
|
 |
|
|
|
|
 |
K.Inaba
(2010).
Structural basis of protein disulfide bond generation in the cell.
|
| |
Genes Cells,
15,
935-943.
|
 |
|
|
|
|
 |
M.Hakim,
and
D.Fass
(2010).
Cytosolic disulfide bond formation in cells infected with large nucleocytoplasmic DNA viruses.
|
| |
Antioxid Redox Signal,
13,
1261-1271.
|
 |
|
|
|
|
 |
N.Heldman,
O.Vonshak,
C.S.Sevier,
E.Vitu,
T.Mehlman,
and
D.Fass
(2010).
Steps in reductive activation of the disulfide-generating enzyme Ero1p.
|
| |
Protein Sci,
19,
1863-1876.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Endo,
K.Yamano,
and
S.Kawano
(2010).
Structural basis for the disulfide relay system in the mitochondrial intermembrane space.
|
| |
Antioxid Redox Signal,
13,
1359-1373.
|
 |
|
|
|
|
 |
V.K.Kodali,
and
C.Thorpe
(2010).
Quiescin sulfhydryl oxidase from Trypanosoma brucei: catalytic activity and mechanism of a QSOX family member with a single thioredoxin domain.
|
| |
Biochemistry,
49,
2075-2085.
|
 |
|
|
|
|
 |
V.K.Kodali,
and
C.Thorpe
(2010).
Oxidative protein folding and the Quiescin-sulfhydryl oxidase family of flavoproteins.
|
| |
Antioxid Redox Signal,
13,
1217-1230.
|
 |
|
|
|
|
 |
W.Li,
S.Schulman,
R.J.Dutton,
D.Boyd,
J.Beckwith,
and
T.A.Rapoport
(2010).
Structure of a bacterial homologue of vitamin K epoxide reductase.
|
| |
Nature,
463,
507-512.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Chacinska,
C.M.Koehler,
D.Milenkovic,
T.Lithgow,
and
N.Pfanner
(2009).
Importing mitochondrial proteins: machineries and mechanisms.
|
| |
Cell,
138,
628-644.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
I.Peluso,
F.Fuccio,
and
M.Porcelli
(2009).
Purine nucleoside phosphorylases from hyperthermophilic Archaea require a CXC motif for stability and folding.
|
| |
FEBS J,
276,
5799-5805.
|
 |
|
|
|
|
 |
S.K.Ang,
and
H.Lu
(2009).
Deciphering structural and functional roles of individual disulfide bonds of the mitochondrial sulfhydryl oxidase Erv1p.
|
| |
J Biol Chem,
284,
28754-28761.
|
 |
|
|
|
|
 |
U.Derewenda,
T.Boczek,
K.L.Gorres,
M.Yu,
L.W.Hung,
D.Cooper,
A.Joachimiak,
R.T.Raines,
and
Z.S.Derewenda
(2009).
Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif .
|
| |
Biochemistry,
48,
8664-8671.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.N.Daithankar,
S.R.Farrell,
and
C.Thorpe
(2009).
Augmenter of liver regeneration: substrate specificity of a flavin-dependent oxidoreductase from the mitochondrial intermembrane space.
|
| |
Biochemistry,
48,
4828-4837.
|
 |
|
|
|
|
 |
D.Sela,
N.Yaffe,
and
J.Shlomai
(2008).
Enzymatic Mechanism Controls Redox-mediated Protein-DNA Interactions at the Replication Origin of Kinetoplast DNA Minicircles.
|
| |
J Biol Chem,
283,
32034-32044.
|
 |
|
|
|
|
 |
D.Stojanovski,
D.Milenkovic,
J.M.Müller,
K.Gabriel,
A.Schulze-Specking,
M.J.Baker,
M.T.Ryan,
B.Guiard,
N.Pfanner,
and
A.Chacinska
(2008).
Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase.
|
| |
J Cell Biol,
183,
195-202.
|
 |
|
|
|
|
 |
E.J.Heckler,
P.C.Rancy,
V.K.Kodali,
and
C.Thorpe
(2008).
Generating disulfides with the Quiescin-sulfhydryl oxidases.
|
| |
Biochim Biophys Acta,
1783,
567-577.
|
 |
|
|
|
|
 |
E.Pedone,
D.Limauro,
and
S.Bartolucci
(2008).
The machinery for oxidative protein folding in thermophiles.
|
| |
Antioxid Redox Signal,
10,
157-170.
|
 |
|
|
|
|
 |
G.Launay,
and
T.Simonson
(2008).
Homology modelling of protein-protein complexes: a simple method and its possibilities and limitations.
|
| |
BMC Bioinformatics,
9,
427.
|
 |
|
|
|
|
 |
M.Mariappan,
S.L.Gande,
K.Radhakrishnan,
B.Schmidt,
T.Dierks,
and
K.von Figura
(2008).
The non-catalytic N-terminal extension of formylglycine-generating enzyme is required for its biological activity and retention in the endoplasmic reticulum.
|
| |
J Biol Chem,
283,
11556-11564.
|
 |
|
|
|
|
 |
B.Heras,
M.Kurz,
S.R.Shouldice,
and
J.L.Martin
(2007).
The name's bond......disulfide bond.
|
| |
Curr Opin Struct Biol,
17,
691-698.
|
 |
|
|
|
|
 |
C.Thorpe,
and
D.L.Coppock
(2007).
Generating disulfides in multicellular organisms: emerging roles for a new flavoprotein family.
|
| |
J Biol Chem,
282,
13929-13933.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
S.Gorassini,
M.F.Mazzeo,
R.A.Siciliano,
V.Carbone,
V.Zappia,
and
M.Porcelli
(2007).
Biochemical and structural characterization of mammalian-like purine nucleoside phosphorylase from the Archaeon Pyrococcus furiosus.
|
| |
FEBS J,
274,
2482-2495.
|
 |
|
|
|
|
 |
J.M.Herrmann,
and
R.Köhl
(2007).
Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria.
|
| |
J Cell Biol,
176,
559-563.
|
 |
|
|
|
|
 |
N.Y.Marcus,
R.A.Marcus,
B.Z.Schmidt,
and
D.B.Haslam
(2007).
Contribution of the HEDJ/ERdj3 cysteine-rich domain to substrate interactions.
|
| |
Arch Biochem Biophys,
468,
147-158.
|
 |
|
|
|
|
 |
O.Dmitrenko,
C.Thorpe,
and
R.D.Bach
(2007).
Mechanism of SN2 disulfide bond cleavage by phosphorus nucleophiles. Implications for biochemical disulfide reducing agents.
|
| |
J Org Chem,
72,
8298-8307.
|
 |
|
|
|
|
 |
W.Wang,
J.R.Winther,
and
C.Thorpe
(2007).
Erv2p: characterization of the redox behavior of a yeast sulfhydryl oxidase.
|
| |
Biochemistry,
46,
3246-3254.
|
 |
|
|
|
|
 |
A.Görlach,
P.Klappa,
and
T.Kietzmann
(2006).
The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control.
|
| |
Antioxid Redox Signal,
8,
1391-1418.
|
 |
|
|
|
|
 |
C.S.Sevier,
and
C.A.Kaiser
(2006).
Disulfide transfer between two conserved cysteine pairs imparts selectivity to protein oxidation by Ero1.
|
| |
Mol Biol Cell,
17,
2256-2266.
|
 |
|
|
|
|
 |
C.S.Sevier,
and
C.A.Kaiser
(2006).
Conservation and diversity of the cellular disulfide bond formation pathways.
|
| |
Antioxid Redox Signal,
8,
797-811.
|
 |
|
|
|
|
 |
C.W.Gruber,
M.Cemazar,
B.Heras,
J.L.Martin,
and
D.J.Craik
(2006).
Protein disulfide isomerase: the structure of oxidative folding.
|
| |
Trends Biochem Sci,
31,
455-464.
|
 |
|
|
|
|
 |
D.L.Coppock,
and
C.Thorpe
(2006).
Multidomain flavin-dependent sulfhydryl oxidases.
|
| |
Antioxid Redox Signal,
8,
300-311.
|
 |
|
|
|
|
 |
E.Gross,
C.S.Sevier,
N.Heldman,
E.Vitu,
M.Bentzur,
C.A.Kaiser,
C.Thorpe,
and
D.Fass
(2006).
Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p.
|
| |
Proc Natl Acad Sci U S A,
103,
299-304.
|
 |
|
|
|
|
 |
I.Rodríguez,
M.Redrejo-Rodríguez,
J.M.Rodríguez,
A.Alejo,
J.Salas,
and
M.L.Salas
(2006).
African swine fever virus pB119L protein is a flavin adenine dinucleotide-linked sulfhydryl oxidase.
|
| |
J Virol,
80,
3157-3166.
|
 |
|
|
|
|
 |
J.D.Rand,
and
C.M.Grant
(2006).
The thioredoxin system protects ribosomes against stress-induced aggregation.
|
| |
Mol Biol Cell,
17,
387-401.
|
 |
|
|
|
|
 |
J.E.Friedman,
J.A.Watson,
D.W.Lam,
and
S.E.Rokita
(2006).
Iodotyrosine deiodinase is the first mammalian member of the NADH oxidase/flavin reductase superfamily.
|
| |
J Biol Chem,
281,
2812-2819.
|
 |
|
|
|
|
 |
J.Kruusma,
A.M.Benham,
J.A.Williams,
and
R.Kataky
(2006).
An introduction to thiol redox proteins in the endoplasmic reticulum and a review of current electrochemical methods of detection of thiols.
|
| |
Analyst,
131,
459-473.
|
 |
|
|
|
|
 |
K.Inaba,
Y.H.Takahashi,
K.Ito,
and
S.Hayashi
(2006).
Critical role of a thiolate-quinone charge transfer complex and its adduct form in de novo disulfide bond generation by DsbB.
|
| |
Proc Natl Acad Sci U S A,
103,
287-292.
|
 |
|
|
|
|
 |
R.Pawlowski,
and
J.Jura
(2006).
ALR and liver regeneration.
|
| |
Mol Cell Biochem,
288,
159-169.
|
 |
|
|
|
|
 |
Y.Furukawa,
and
T.V.O'Halloran
(2006).
Posttranslational modifications in Cu,Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis.
|
| |
Antioxid Redox Signal,
8,
847-867.
|
 |
|
|
|
|
 |
A.Preusser-Kunze,
M.Mariappan,
B.Schmidt,
S.L.Gande,
K.Mutenda,
D.Wenzel,
K.von Figura,
and
T.Dierks
(2005).
Molecular characterization of the human Calpha-formylglycine-generating enzyme.
|
| |
J Biol Chem,
280,
14900-14910.
|
 |
|
|
|
|
 |
C.S.Sevier,
H.Kadokura,
V.C.Tam,
J.Beckwith,
D.Fass,
and
C.A.Kaiser
(2005).
The prokaryotic enzyme DsbB may share key structural features with eukaryotic disulfide bond forming oxidoreductases.
|
| |
Protein Sci,
14,
1630-1642.
|
 |
|
|
|
|
 |
E.van Anken,
and
I.Braakman
(2005).
Versatility of the endoplasmic reticulum protein folding factory.
|
| |
Crit Rev Biochem Mol Biol,
40,
191-228.
|
 |
|
|
|
|
 |
G.Cacciapuoti,
S.Forte,
M.A.Moretti,
A.Brio,
V.Zappia,
and
M.Porcelli
(2005).
A novel hyperthermostable 5'-deoxy-5'-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus.
|
| |
FEBS J,
272,
1886-1899.
|
 |
|
|
|
|
 |
N.Mesecke,
N.Terziyska,
C.Kozany,
F.Baumann,
W.Neupert,
K.Hell,
and
J.M.Herrmann
(2005).
A disulfide relay system in the intermembrane space of mitochondria that mediates protein import.
|
| |
Cell,
121,
1059-1069.
|
 |
|
|
|
|
 |
A.Levitan,
A.Danon,
and
T.Lisowsky
(2004).
Unique features of plant mitochondrial sulfhydryl oxidase.
|
| |
J Biol Chem,
279,
20002-20008.
|
 |
|
|
|
|
 |
B.P.Tu,
and
J.S.Weissman
(2004).
Oxidative protein folding in eukaryotes: mechanisms and consequences.
|
| |
J Cell Biol,
164,
341-346.
|
 |
|
|
|
|
 |
E.Gross,
D.B.Kastner,
C.A.Kaiser,
and
D.Fass
(2004).
Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell.
|
| |
Cell,
117,
601-610.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Cacciapuoti,
M.A.Moretti,
S.Forte,
A.Brio,
L.Camardella,
V.Zappia,
and
M.Porcelli
(2004).
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and assignment of disulfide bonds.
|
| |
Eur J Biochem,
271,
4834-4844.
|
 |
|
|
|
|
 |
G.T.Hanson,
R.Aggeler,
D.Oglesbee,
M.Cannon,
R.A.Capaldi,
R.Y.Tsien,
and
S.J.Remington
(2004).
Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
|
| |
J Biol Chem,
279,
13044-13053.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.C.Bardwell
(2004).
The dance of disulfide formation.
|
| |
Nat Struct Mol Biol,
11,
582-583.
|
 |
|
|
|
|
 |
M.Varsányi,
A.Szarka,
E.Papp,
D.Makai,
G.Nardai,
R.Fulceri,
P.Csermely,
J.Mandl,
A.Benedetti,
and
G.Bánhegyi
(2004).
FAD transport and FAD-dependent protein thiol oxidation in rat liver microsomes.
|
| |
J Biol Chem,
279,
3370-3374.
|
 |
|
|
|
|
 |
R.Bass,
L.W.Ruddock,
P.Klappa,
and
R.B.Freedman
(2004).
A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein.
|
| |
J Biol Chem,
279,
5257-5262.
|
 |
|
|
|
|
 |
A.K.Galande,
J.O.Trent,
and
A.F.Spatola
(2003).
Understanding base-assisted desulfurization using a variety of disulfide-bridged peptides.
|
| |
Biopolymers,
71,
534-551.
|
 |
|
|
|
|
 |
C.K.Wu,
T.A.Dailey,
H.A.Dailey,
B.C.Wang,
and
J.P.Rose
(2003).
The crystal structure of augmenter of liver regeneration: A mammalian FAD-dependent sulfhydryl oxidase.
|
| |
Protein Sci,
12,
1109-1118.
|
 |
|
|
|
|
 |
G.Bánhegyi,
M.Csala,
A.Szarka,
M.Varsányi,
A.Benedetti,
and
J.Mandl
(2003).
Role of ascorbate in oxidative protein folding.
|
| |
Biofactors,
17,
37-46.
|
 |
|
|
|
|
 |
G.Hofhaus,
J.E.Lee,
I.Tews,
B.Rosenberg,
and
T.Lisowsky
(2003).
The N-terminal cysteine pair of yeast sulfhydryl oxidase Erv1p is essential for in vivo activity and interacts with the primary redox centre.
|
| |
Eur J Biochem,
270,
1528-1535.
|
 |
|
|
|
|
 |
K.J.Woycechowsky,
and
R.T.Raines
(2003).
The CXC motif: a functional mimic of protein disulfide isomerase.
|
| |
Biochemistry,
42,
5387-5394.
|
 |
|
|
|
|
 |
P.Tarnow,
T.Schoneberg,
H.Krude,
A.Gruters,
and
H.Biebermann
(2003).
Mutationally induced disulfide bond formation within the third extracellular loop causes melanocortin 4 receptor inactivation in patients with obesity.
|
| |
J Biol Chem,
278,
48666-48673.
|
 |
|
|
|
|
 |
X.Chen,
Y.Li,
K.Wei,
L.Li,
W.Liu,
Y.Zhu,
Z.Qiu,
and
F.He
(2003).
The potentiation role of hepatopoietin on activator protein-1 is dependent on its sulfhydryl oxidase activity.
|
| |
J Biol Chem,
278,
49022-49030.
|
 |
|
|
|
|
 |
B.P.Tu,
and
J.S.Weissman
(2002).
The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum.
|
| |
Mol Cell,
10,
983-994.
|
 |
|
|
|
|
 |
J.F.Collet,
and
J.C.Bardwell
(2002).
Disulfides out of thin air.
|
| |
Nat Struct Biol,
9,
2-3.
|
 |
|
|
|
|
 |
J.Messens,
J.C.Martins,
K.Van Belle,
E.Brosens,
A.Desmyter,
M.De Gieter,
J.M.Wieruszeski,
R.Willem,
L.Wyns,
and
I.Zegers
(2002).
All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
|
| |
Proc Natl Acad Sci U S A,
99,
8506-8511.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.G.Senkevich,
C.L.White,
E.V.Koonin,
and
B.Moss
(2002).
Complete pathway for protein disulfide bond formation encoded by poxviruses.
|
| |
Proc Natl Acad Sci U S A,
99,
6667-6672.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

