 |
PDBsum entry 1jqi

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1jqi
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of rat short chain acyl-coa dehydrogenase complexed with acetoacetyl-coa
|
|
Structure:
|
 |
Short chain acyl-coa dehydrogenase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Rattus norvegicus. Norway rat. Organism_taxid: 10116. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.168
|
R-free:
|
0.206
|
|
|
Authors:
|
 |
K.P.Battaile,J.Molin-Case,R.Paschke,M.Wang,D.Bennett,J.Vockley,J.- J.P.Kim
|
Key ref:

|
 |
K.P.Battaile
et al.
(2002).
Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
J Biol Chem,
277,
12200-12207.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-Aug-01
|
Release date:
|
13-Feb-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15651
(ACADS_RAT) -
Short-chain specific acyl-CoA dehydrogenase, mitochondrial from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
412 a.a.
384 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.8.1
- short-chain acyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a short-chain 2,3-saturated fatty acyl-CoA + oxidized [electron-transfer flavoprotein] + H+ = a short-chain (2E)-enoyl-CoA + reduced [electron- transfer flavoprotein]
|
 |
 |
 |
 |
 |
Butanoyl-CoA
Bound ligand (Het Group name = )
matches with 98.15% similarity
|
+
|
electron-transfer flavoprotein
|
=
|
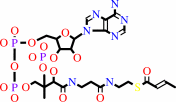 2-butenoyl-CoA
2-butenoyl-CoA
|
+
|
reduced electron-transfer flavoprotein
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:12200-12207
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
|
|
K.P.Battaile,
J.Molin-Case,
R.Paschke,
M.Wang,
D.Bennett,
J.Vockley,
J.J.Kim.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The acyl-CoA dehydrogenases are a family of flavin adenine
dinucleotide-containing enzymes that catalyze the first step in the
beta-oxidation of fatty acids and catabolism of some amino acids. They exhibit
high sequence identity and yet are quite specific in their substrate binding.
Short chain acyl-CoA dehydrogenase has maximal activity toward butyryl-CoA and
negligible activity toward substrates longer than octanoyl-CoA. The crystal
structure of rat short chain acyl-CoA dehydrogenase complexed with the inhibitor
acetoacetyl-CoA has been determined at 2.25 A resolution. Short chain acyl-CoA
dehydrogenase is a homotetramer with a subunit mass of 43 kDa and crystallizes
in the space group P321 with a = 143.61 A and c = 77.46 A. There are two
monomers in the asymmetric unit. The overall structure of short chain acyl-CoA
dehydrogenase is very similar to those of medium chain acyl-CoA dehydrogenase,
isovaleryl-CoA dehydrogenase, and bacterial short chain acyl-CoA dehydrogenase
with a three-domain structure composed of N- and C-terminal alpha-helical
domains separated by a beta-sheet domain. Comparison to other acyl-CoA
dehydrogenases has provided additional insight into the basis of substrate
specificity and the nature of the oxidase activity in this enzyme family. Ten
reported pathogenic human mutations and two polymorphisms have been mapped onto
the structure of short chain acyl-CoA dehydrogenase. None of the mutations
directly affect the binding cavity or intersubunit interactions.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. The overall polypeptide folding of the SCAD
monomer. A, ribbon diagram of the SCAD monomer. Helices are
labeled A-K and  -strands
1-7. FAD is rendered in yellow and acetoacetyl-CoA in green. B,
stereo view of overlay of SCAD (blue), MCAD (red), bSCAD
(green), and IVD (gray). FAD is rendered in yellow and
acetoacetyl-CoA in indigo. Orientation of the monomer is the
same as in A. The figure was generated using Molscript (44) and
Raster3D (45). -strands
1-7. FAD is rendered in yellow and acetoacetyl-CoA in green. B,
stereo view of overlay of SCAD (blue), MCAD (red), bSCAD
(green), and IVD (gray). FAD is rendered in yellow and
acetoacetyl-CoA in indigo. Orientation of the monomer is the
same as in A. The figure was generated using Molscript (44) and
Raster3D (45).
|
 |
Figure 3.
Fig. 3. Stereo diagram of acetoacetyl-CoA and amino acid
residues involved in binding the CoA moiety in SCAD.
Acetoacetyl-CoA is rendered in ball-and-stick format, and amino
acids are rendered as sticks. The acetoacetyl-CoA is "outside"
and all amino acids are "inside" of the molecular surface of the
enzyme. The molecular surface was generated with a 1.4-Å
probe using the program Grasp (46). Molscript (44) and Raster3D
(45) were used to render the image.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
12200-12207)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.J.Erb,
G.Fuchs,
and
B.E.Alber
(2009).
(2S)-Methylsuccinyl-CoA dehydrogenase closes the ethylmalonyl-CoA pathway for acetyl-CoA assimilation.
|
| |
Mol Microbiol,
73,
992.
|
 |
|
|
|
|
 |
Z.Swigonová,
A.W.Mohsen,
and
J.Vockley
(2009).
Acyl-CoA dehydrogenases: Dynamic history of protein family evolution.
|
| |
J Mol Evol,
69,
176-193.
|
 |
|
|
|
|
 |
J.I.Yeh,
U.Chinte,
and
S.Du
(2008).
Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
|
| |
Proc Natl Acad Sci U S A,
105,
3280-3285.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.A.Maggio-Hall,
P.Lyne,
J.A.Wolff,
and
N.P.Keller
(2008).
A single acyl-CoA dehydrogenase is required for catabolism of isoleucine, valine and short-chain fatty acids in Aspergillus nidulans.
|
| |
Fungal Genet Biol,
45,
180-189.
|
 |
|
|
|
|
 |
R.Jethva,
M.J.Bennett,
and
J.Vockley
(2008).
Short-chain acyl-coenzyme A dehydrogenase deficiency.
|
| |
Mol Genet Metab,
95,
195-200.
|
 |
|
|
|
|
 |
R.P.McAndrew,
Y.Wang,
A.W.Mohsen,
M.He,
J.Vockley,
and
J.J.Kim
(2008).
Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase.
|
| |
J Biol Chem,
283,
9435-9443.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.S.Goetzman,
Y.Wang,
M.He,
A.W.Mohsen,
B.K.Ninness,
and
J.Vockley
(2007).
Expression and characterization of mutations in human very long-chain acyl-CoA dehydrogenase using a prokaryotic system.
|
| |
Mol Genet Metab,
91,
138-147.
|
 |
|
|
|
|
 |
S.Gobin-Limballe,
F.Djouadi,
F.Aubey,
S.Olpin,
B.S.Andresen,
S.Yamaguchi,
H.Mandel,
T.Fukao,
J.P.Ruiter,
R.J.Wanders,
R.McAndrew,
J.J.Kim,
and
J.Bastin
(2007).
Genetic basis for correction of very-long-chain acyl-coenzyme A dehydrogenase deficiency by bezafibrate in patient fibroblasts: toward a genotype-based therapy.
|
| |
Am J Hum Genet,
81,
1133-1143.
|
 |
|
|
|
|
 |
D.Binns,
T.Januszewski,
Y.Chen,
J.Hill,
V.S.Markin,
Y.Zhao,
C.Gilpin,
K.D.Chapman,
R.G.Anderson,
and
J.M.Goodman
(2006).
An intimate collaboration between peroxisomes and lipid bodies.
|
| |
J Cell Biol,
173,
719-731.
|
 |
|
|
|
|
 |
L.P.O'Reilly,
B.S.Andresen,
and
P.C.Engel
(2005).
Two novel variants of human medium chain acyl-CoA dehydrogenase (MCAD). K364R, a folding mutation, and R256T, a catalytic-site mutation resulting in a well-folded but totally inactive protein.
|
| |
FEBS J,
272,
4549-4557.
|
 |
|
|
|
|
 |
S.Bhattacharyya,
S.Ma,
M.T.Stankovich,
D.G.Truhlar,
and
J.Gao
(2005).
Potential of mean force calculation for the proton and hydride transfer reactions catalyzed by medium-chain acyl-CoA dehydrogenase: effect of mutations on enzyme catalysis.
|
| |
Biochemistry,
44,
16549-16562.
|
 |
|
|
|
|
 |
A.Nagpal,
M.P.Valley,
P.F.Fitzpatrick,
and
A.M.Orville
(2004).
Crystallization and preliminary analysis of active nitroalkane oxidase in three crystal forms.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1456-1460.
|
 |
|
|
|
|
 |
J.J.Kim,
and
R.Miura
(2004).
Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences.
|
| |
Eur J Biochem,
271,
483-493.
|
 |
|
|
|
|
 |
L.Pedersen,
and
A.Henriksen
(2004).
Expression, purification and crystallization of two peroxisomal acyl-CoA oxidases from Arabidopsis thaliana.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1125-1128.
|
 |
|
|
|
|
 |
N.Gregersen,
P.Bross,
and
B.S.Andresen
(2004).
Genetic defects in fatty acid beta-oxidation and acyl-CoA dehydrogenases. Molecular pathogenesis and genotype-phenotype relationships.
|
| |
Eur J Biochem,
271,
470-482.
|
 |
|
|
|
|
 |
M.G.Thomas,
Y.A.Chan,
and
S.G.Ozanick
(2003).
Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster.
|
| |
Antimicrob Agents Chemother,
47,
2823-2830.
|
 |
|
|
|
|
 |
T.V.Nguyen,
C.Riggs,
D.Babovic-Vuksanovic,
Y.S.Kim,
J.F.Carpenter,
T.P.Burghardt,
N.Gregersen,
and
J.Vockley
(2002).
Purification and characterization of two polymorphic variants of short chain acyl-CoA dehydrogenase reveal reduction of catalytic activity and stability of the Gly185Ser enzyme.
|
| |
Biochemistry,
41,
11126-11133.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

