 |
PDBsum entry 1j9z

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1j9z
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.6.2.4
- NADPH--hemoprotein reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 oxidized [cytochrome P450] + NADPH = 2 reduced [cytochrome P450] + NADP+ + H+
|
 |
 |
 |
 |
 |
2
×
oxidized [cytochrome P450]
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADPH
NADPH
|
=
|
2
×
reduced [cytochrome P450]
|
+
|
 NADP(+)
NADP(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; FMN
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
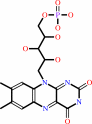 FMN
FMN
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
276:29163-29170
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
|
|
P.A.Hubbard,
A.L.Shen,
R.Paschke,
C.B.Kasper,
J.J.Kim.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
NADPH-cytochrome P450 oxidoreductase catalyzes transfer of electrons from NADPH,
via two flavin cofactors, to various cytochrome P450s. The crystal structure of
the rat reductase complexed with NADP(+) has revealed that nicotinamide access
to FAD is blocked by an aromatic residue (Trp-677), which stacks against the
re-face of the isoalloxazine ring of the flavin. To investigate the nature of
interactions between the nicotinamide, FAD, and Trp-677 during the catalytic
cycle, three mutant proteins were studied by crystallography. The first mutant,
W677X, has the last two C-terminal residues, Trp-677 and Ser-678, removed; the
second mutant, W677G, retains the C-terminal serine residue. The third mutant
has the following three catalytic residues substituted: S457A, C630A, and D675N.
In the W677X and W677G structures, the nicotinamide moiety of NADP(+) lies
against the FAD isoalloxazine ring with a tilt of approximately 30 degrees
between the planes of the two rings. These results, together with the
S457A/C630A/D675N structure, allow us to propose a mechanism for hydride
transfer regulated by changes in hydrogen bonding and pi-pi interactions between
the isoalloxazine ring and either the nicotinamide ring or Trp-677 indole ring.
Superimposition of the mutant and wild-type structures shows significant
mobility between the two flavin domains of the enzyme. This, together with the
high degree of disorder observed in the FMN domain of all three mutant
structures, suggests that conformational changes occur during catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Stereo diagram of an F[o]  F[c] OMIT
map contoured at 2 F[c] OMIT
map contoured at 2  from the
W667X deletion mutant. The density shows the A-face of the
nicotinamide ring of NADP+ to stack against the re-face of the
isoalloxazine ring of FAD, with a ~30° tilt between the
planes of the two rings. from the
W667X deletion mutant. The density shows the A-face of the
nicotinamide ring of NADP+ to stack against the re-face of the
isoalloxazine ring of FAD, with a ~30° tilt between the
planes of the two rings.
|
 |
Figure 4.
Fig. 4. Diagram showing the hydrogen bond networks
surrounding the isoalloxazine ring of FAD and the nicotinamide
ring of NADP+. Thin dashed lines represent hydrogen bonds; thick
dotted lines are van der Waal's contacts. A, the wild-type
structure. The two catalytic waters involved in hydrogen bond
networks with FAD are included. B, the W677X mutant structure.
The presence of the nicotinamide ring results in Asp-675 moving
out of hydrogen bonding distance from Ser-457 and Cys-630. Van
der Waal's contacts are formed between the C-4 atom of the
nicotinamide ring and Cys-630 and from Ser-457 to the N-5 atom
of FAD. Another van der Waal's contact is formed between the C-4
atom of the nicotinamide ring and the N-5 atom of FAD.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2001,
276,
29163-29170)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Degregorio,
S.J.Sadeghi,
G.Di Nardo,
G.Gilardi,
and
S.P.Solinas
(2011).
Understanding uncoupling in the multiredox centre P450 3A4-BMR model system.
|
| |
J Biol Inorg Chem,
16,
109-116.
|
 |
|
|
|
|
 |
D.Sandee,
K.Morrissey,
V.Agrawal,
H.K.Tam,
M.A.Kramer,
T.S.Tracy,
K.M.Giacomini,
and
W.L.Miller
(2010).
Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro.
|
| |
Pharmacogenet Genomics,
20,
677-686.
|
 |
|
|
|
|
 |
V.Agrawal,
J.H.Choi,
K.M.Giacomini,
and
W.L.Miller
(2010).
Substrate-specific modulation of CYP3A4 activity by genetic variants of cytochrome P450 oxidoreductase.
|
| |
Pharmacogenet Genomics,
20,
611-618.
|
 |
|
|
|
|
 |
C.Xia,
I.Misra,
T.Iyanagi,
and
J.J.Kim
(2009).
Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase.
|
| |
J Biol Chem,
284,
30708-30717.
|
 |
|
|
|
|
 |
D.Hamdane,
C.Xia,
S.C.Im,
H.Zhang,
J.J.Kim,
and
L.Waskell
(2009).
Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450.
|
| |
J Biol Chem,
284,
11374-11384.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Beaumont,
J.C.Lambry,
M.Blanchard-Desce,
P.Martasek,
S.P.Panda,
E.E.van Faassen,
J.C.Brochon,
E.Deprez,
and
A.Slama-Schwok
(2009).
NO formation by neuronal NO-synthase can be controlled by ultrafast electron injection from a nanotrigger.
|
| |
Chembiochem,
10,
690-701.
|
 |
|
|
|
|
 |
J.Ellis,
A.Gutierrez,
I.L.Barsukov,
W.C.Huang,
J.G.Grossmann,
and
G.C.Roberts
(2009).
Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering.
|
| |
J Biol Chem,
284,
36628-36637.
|
 |
|
|
|
|
 |
R.P.Ilagan,
M.Tiso,
D.W.Konas,
C.Hemann,
D.Durra,
R.Hille,
and
D.J.Stuehr
(2008).
Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins.
|
| |
J Biol Chem,
283,
19603-19615.
|
 |
|
|
|
|
 |
S.N.Hart,
and
X.B.Zhong
(2008).
P450 oxidoreductase: genetic polymorphisms and implications for drug metabolism and toxicity.
|
| |
Expert Opin Drug Metab Toxicol,
4,
439-452.
|
 |
|
|
|
|
 |
V.Agrawal,
N.Huang,
and
W.L.Miller
(2008).
Pharmacogenetics of P450 oxidoreductase: effect of sequence variants on activities of CYP1A2 and CYP2C19.
|
| |
Pharmacogenet Genomics,
18,
569-576.
|
 |
|
|
|
|
 |
A.W.Munro,
H.M.Girvan,
and
K.J.McLean
(2007).
Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily.
|
| |
Nat Prod Rep,
24,
585-609.
|
 |
|
|
|
|
 |
C.E.Flück,
C.Nicolo,
and
A.V.Pandey
(2007).
Clinical, structural and functional implications of mutations and polymorphisms in human NADPH P450 oxidoreductase.
|
| |
Fundam Clin Pharmacol,
21,
399-410.
|
 |
|
|
|
|
 |
K.J.McLean,
H.M.Girvan,
and
A.W.Munro
(2007).
Cytochrome P450/redox partner fusion enzymes: biotechnological and toxicological prospects.
|
| |
Expert Opin Drug Metab Toxicol,
3,
847-863.
|
 |
|
|
|
|
 |
J.Hritz,
G.Zoldák,
and
E.Sedlák
(2006).
Cofactor assisted gating mechanism in the active site of NADH oxidase from Thermus thermophilus.
|
| |
Proteins,
64,
465-476.
|
 |
|
|
|
|
 |
M.Jáchymová,
P.Martásek,
S.Panda,
L.J.Roman,
M.Panda,
T.M.Shea,
Y.Ishimura,
J.J.Kim,
and
B.S.Masters
(2005).
Recruitment of governing elements for electron transfer in the nitric oxide synthase family.
|
| |
Proc Natl Acad Sci U S A,
102,
15833-15838.
|
 |
|
|
|
|
 |
P.Meinhold,
M.W.Peters,
M.M.Chen,
K.Takahashi,
and
F.H.Arnold
(2005).
Direct conversion of ethane to ethanol by engineered cytochrome P450 BM3.
|
| |
Chembiochem,
6,
1765-1768.
|
 |
|
|
|
|
 |
A.J.Dunford,
K.R.Marshall,
A.W.Munro,
and
N.S.Scrutton
(2004).
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in the region of the FAD shielding residue Phe1395.
|
| |
Eur J Biochem,
271,
2548-2560.
|
 |
|
|
|
|
 |
A.Gutierrez,
A.W.Munro,
A.Grunau,
C.R.Wolf,
N.S.Scrutton,
and
G.C.Roberts
(2003).
Interflavin electron transfer in human cytochrome P450 reductase is enhanced by coenzyme binding. Relaxation kinetic studies with coenzyme analogues.
|
| |
Eur J Biochem,
270,
2612-2621.
|
 |
|
|
|
|
 |
D.Leys,
J.Basran,
F.Talfournier,
M.J.Sutcliffe,
and
N.S.Scrutton
(2003).
Extensive conformational sampling in a ternary electron transfer complex.
|
| |
Nat Struct Biol,
10,
219-225.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Zoldák,
R.Sut'ák,
M.Antalík,
M.Sprinzl,
and
E.Sedlák
(2003).
Role of conformational flexibility for enzymatic activity in NADH oxidase from Thermus thermophilus.
|
| |
Eur J Biochem,
270,
4887-4897.
|
 |
|
|
|
|
 |
R.D.Finn,
J.Basran,
O.Roitel,
C.R.Wolf,
A.W.Munro,
M.J.Paine,
and
N.S.Scrutton
(2003).
Determination of the redox potentials and electron transfer properties of the FAD- and FMN-binding domains of the human oxidoreductase NR1.
|
| |
Eur J Biochem,
270,
1164-1175.
|
 |
|
|
|
|
 |
S.Adak,
M.Sharma,
A.L.Meade,
and
D.J.Stuehr
(2002).
A conserved flavin-shielding residue regulates NO synthase electron transfer and nicotinamide coenzyme specificity.
|
| |
Proc Natl Acad Sci U S A,
99,
13516-13521.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

