 |
PDBsum entry 1hye

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1hye
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.375
- L-2-hydroxycarboxylate dehydrogenase [NAD(P)(+)].
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a (2S)-2-hydroxycarboxylate + NAD+ = a 2-oxocarboxylate + NADH + H+
|
|
2.
|
a (2S)-2-hydroxycarboxylate + NADP+ = a 2-oxocarboxylate + NADPH + H+
|
|
 |
 |
 |
 |
 |
(2S)-2-hydroxycarboxylate
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
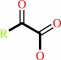 2-oxocarboxylate
2-oxocarboxylate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
(2S)-2-hydroxycarboxylate
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 2-oxocarboxylate
2-oxocarboxylate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
307:1351-1362
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the MJ0490 gene product of the hyperthermophilic archaebacterium Methanococcus jannaschii, a novel member of the lactate/malate family of dehydrogenases.
|
|
B.I.Lee,
C.Chang,
S.J.Cho,
S.H.Eom,
K.K.Kim,
Y.G.Yu,
S.W.Suh.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The MJ0490 gene, one of the only two genes of Methanococcus jannaschii showing
sequence similarity to the lactate/malate family of dehydrogenases, was
classified initially as coding for a putative l-lactate dehydrogenase (LDH). It
has been re-classified as a malate dehydrogenase (MDH) gene, because it shows
significant sequence similarity to MT0188, MDH II from Methanobacterium
thermoautotrophicum strain DeltaH. The three-dimensional structure of its gene
product has been determined in two crystal forms: a "dimeric"
structure in the orthorhombic crystal at 1.9 A resolution and a
"tetrameric" structure in the tetragonal crystal at 2.8 A. These
structures share a similar subunit fold with other LDHs and MDHs. The tetrameric
structure resembles typical tetrameric LDHs. The dimeric structure is equivalent
to the P-dimer of tetrameric LDHs, unlike dimeric MDHs, which correspond to the
Q-dimer. The structure reveals that the cofactor NADP(H) is bound at the active
site, despite the fact that it was not intentionally added during protein
purification and crystallization. The preference of NADP(H) over NAD(H) has been
supported by activity assays. The cofactor preference is explained by the
presence of a glycine residue in the cofactor binding pocket (Gly33), which
replaces a conserved aspartate (or glutamate) residue in other NAD-dependent
LDHs or MDHs. Preference for NADP(H) is contributed by hydrogen bonds between
the oxygen atoms of the monophosphate group and the ribose sugar of adenosine in
NADP(H) and the side-chains of Ser9, Arg34, His36, and Ser37. The MDH activity
of MJ0490 is made possible by Arg86, which is conserved in MDHs but not in LDHs.
The enzymatic assay showed that the MJ0490 protein possesses the
fructose-1,6-bisphosphate-activated LDH activity (reduction). Thus the MJ0490
gene product appears to be a novel member of the lactate/malate dehydrogenase
family, displaying an LDH scaffold and exhibiting a relaxed substrate and
cofactor specificities in NADP(H) and NAD(H)-dependent malate and lactate
dehydrogenase reactions.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Overall structure of MJ0490 protein. (a) Stereo
ribbon diagram of the subunit model, and stereo C^a tracings of
(b) terameric structure and (c) dimeric structure. NADP+
molecules are shown in orange (in (a)) or in blue (in (b), and
(c)). These Figures were drawn by MOLSCRIPT [Kraulis 1991] and
Raster3D [Merritt and Murphy 1994].
|
 |
Figure 3.
Figure 3. (a) Stereo view of the final (2F[o] - F[c])
electron density map around the bound NADP+, calculated using
20.0-1.9 Å data and contoured at 1.0s. (b) Stereo view of
the cofactor binding site. The NADP+ molecule is in dark olive
green and interacting residues are drawn in orange. Broken lines
indicate hydrogen bonds or close contacts.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2001,
307,
1351-1362)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.J.Yennaco,
Y.Hu,
and
J.F.Holden
(2007).
Characterization of malate dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
|
| |
Extremophiles,
11,
741-746.
|
 |
|
|
|
|
 |
T.Fujii,
T.Oikawa,
I.Muraoka,
K.Soda,
and
Y.Hata
(2007).
Crystallization and preliminary X-ray diffraction studies of tetrameric malate dehydrogenase from the novel Antarctic psychrophile Flavobacterium frigidimaris KUC-1.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
983-986.
|
 |
|
|
|
|
 |
G.Yang,
C.Jing,
P.Zhu,
X.Hu,
J.Xu,
Z.Wu,
and
X.Yu
(2006).
Molecular cloning and characterization of a novel lactate dehydrogenase gene from Clonorchis sinensis.
|
| |
Parasitol Res,
99,
55-64.
|
 |
|
|
|
|
 |
L.L.Grochowski,
H.Xu,
and
R.H.White
(2006).
Identification of lactaldehyde dehydrogenase in Methanocaldococcus jannaschii and its involvement in production of lactate for F420 biosynthesis.
|
| |
J Bacteriol,
188,
2836-2844.
|
 |
|
|
|
|
 |
M.Tehei,
R.Daniel,
and
G.Zaccai
(2006).
Fundamental and biotechnological applications of neutron scattering measurements for macromolecular dynamics.
|
| |
Eur Biophys J,
35,
551-558.
|
 |
|
|
|
|
 |
M.Tehei,
D.Madern,
B.Franzetti,
and
G.Zaccai
(2005).
Neutron scattering reveals the dynamic basis of protein adaptation to extreme temperature.
|
| |
J Biol Chem,
280,
40974-40979.
|
 |
|
|
|
|
 |
S.Cheek,
Y.Qi,
S.S.Krishna,
L.N.Kinch,
and
N.V.Grishin
(2004).
4SCOPmap: automated assignment of protein structures to evolutionary superfamilies.
|
| |
BMC Bioinformatics,
5,
197.
|
 |
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

