 |
PDBsum entry 1h2t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Nuclear protein
|
PDB id
|
|

|

|
1h2t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Large-Scale induced fit recognition of an m(7)gpppg cap analogue by the human nuclear cap-Binding complex.
|
 |
|
Authors
|
 |
C.Mazza,
A.Segref,
I.W.Mattaj,
S.Cusack.
|
 |
|
Ref.
|
 |
EMBO J, 2002,
21,
5548-5557.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
The heterodimeric nuclear cap-binding complex (CBC) binds to the 5' cap
structure of RNAs in the nucleus and plays a central role in their diverse
maturation steps. We describe the crystal structure at 2.1 A resolution of human
CBC bound to an m(7)GpppG cap analogue. Comparison with the structure of
uncomplexed CBC shows that cap binding induces co-operative folding around the
dinucleotide of some 50 residues from the N- and C-terminal extensions to the
central RNP domain of the small subunit CBP20. The cap-bound conformation of
CBP20 is stabilized by an intricate network of interactions both to the ligand
and within the subunit, as well as new interactions of the CBP20 N-terminal tail
with the large subunit CBP80. Although the structure is very different from that
of other known cap-binding proteins, such as the cytoplasmic cap-binding protein
eIF4E, specificity for the methylated guanosine again is achieved by sandwiching
the base between two aromatic residues, in this case two conserved tyrosines.
Implications for the transfer of capped mRNAs to eIF4E, required for translation
initiation, are discussed.
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 (A) Time course of limited proteolysis. A 42  g
aliquot of CBC was incubated at room temperature with 210 ng of
trypsin in the absence or presence of 10 mM cap analogue
m^7GpppG in a total volume of 60 g
aliquot of CBC was incubated at room temperature with 210 ng of
trypsin in the absence or presence of 10 mM cap analogue
m^7GpppG in a total volume of 60  l.
Aliquots of 10 l.
Aliquots of 10  l
were taken off every 0, 5, 10, 20, 40 and 60 min, denatured by 3 l
were taken off every 0, 5, 10, 20, 40 and 60 min, denatured by 3
 l
of denaturing buffer (125 mM Tris pH 6.8, 260 mM DTT, 30%
glycerol, 10% SDS and 0.025% Coomassie Blue) and loaded onto a
13.5% Tricine SDS−polyacrylamide gel. (B) Cap-binding activity
of CBP80 l
of denaturing buffer (125 mM Tris pH 6.8, 260 mM DTT, 30%
glycerol, 10% SDS and 0.025% Coomassie Blue) and loaded onto a
13.5% Tricine SDS−polyacrylamide gel. (B) Cap-binding activity
of CBP80  653−701.
[^35S]Methionine-labelled wild-type or mutant CBP80 was
incubated for 30 min at room temperature in the absence (−) or
presence of either 1.7 653−701.
[^35S]Methionine-labelled wild-type or mutant CBP80 was
incubated for 30 min at room temperature in the absence (−) or
presence of either 1.7  M
(high) or 53 nM (low) m^7GpppG-capped (m7) or ApppG-capped (A)
unlabelled U1 M
(high) or 53 nM (low) m^7GpppG-capped (m7) or ApppG-capped (A)
unlabelled U1  Sm
RNAs. The samples were fractionated by native 6% PAGE followed
by fluorography. Free CBP80, CBC and the CBC−RNA complex are
indicated. In the lanes indicated by an asterisk, the
corresponding CBP80 proteins were loaded without CBP20. (C)
Shuttling activity of CBP80 Sm
RNAs. The samples were fractionated by native 6% PAGE followed
by fluorography. Free CBP80, CBC and the CBC−RNA complex are
indicated. In the lanes indicated by an asterisk, the
corresponding CBP80 proteins were loaded without CBP20. (C)
Shuttling activity of CBP80  NLS NLS
 653−701
in Xenopus oocytes. [^35S]methionine-labelled CBP80 653−701
in Xenopus oocytes. [^35S]methionine-labelled CBP80  NLS2
or CBP80 NLS2
or CBP80  NLS2 NLS2
 653−701
were injected together with [^35S]methionine-labelled GST−M10
into Xenopus oocyte nuclei either (1) alone (lanes 1 and 2, and
7 and 8), (2) together with m7GpppG-capped unlabelled U1 653−701
were injected together with [^35S]methionine-labelled GST−M10
into Xenopus oocyte nuclei either (1) alone (lanes 1 and 2, and
7 and 8), (2) together with m7GpppG-capped unlabelled U1  Sm
RNAs (lanes 3 and 4, and 9 and 10) or (3) together with
ApppG-capped U1 Sm
RNAs (lanes 3 and 4, and 9 and 10) or (3) together with
ApppG-capped U1  Sm
RNAs (lanes 5 and 6, and 11 and12). Oocytes were dissected
either immediately (lanes 1 and 2, and 7 and 8) or 5 h after
injection (lanes 3−6 and 9−12) and the proteins analysed by
SDS−PAGE followed by fluorography. GST−M10 is a mutant of
HIV Rev with a non-functional nuclear export signal used as a
negative control. See Ohno et al. (2000) or Segref et al. (2001)
for more details. Sm
RNAs (lanes 5 and 6, and 11 and12). Oocytes were dissected
either immediately (lanes 1 and 2, and 7 and 8) or 5 h after
injection (lanes 3−6 and 9−12) and the proteins analysed by
SDS−PAGE followed by fluorography. GST−M10 is a mutant of
HIV Rev with a non-functional nuclear export signal used as a
negative control. See Ohno et al. (2000) or Segref et al. (2001)
for more details.
|
 |
Figure 2.
Figure 2 (A) Structure of CBP20 in the CBC  NLS
complex. Ribbon representation of the cap-free conformation of
CBP20 showing residues 30−125 (grey). Residues involved in cap
binding are shown in yellow (already in their cap-bound
conformation) and blue (undergo a conformational change to
interact with the cap) (compare with B). Phe49 (light green)
changes conformation to help stabilize the C-terminal domain
(compare with B). Salt bridges and hydrogen bonds are indicated
by dashed lines. (B) Stabilization of the N- and C-terminal
extensions of CBP20 upon cap binding. Ribbon representation of
cap-bound CBP20 showing residues 32−125 (already ordered in
the cap-free form) in grey and the N- and C-terminal extensions
that fold upon cap binding in green and orange, respectively.
Yellow residues stabilize the folded conformation of these two
extensions through interactions with the cap. This stabilization
is reinforced by protein−protein interactions involving the
light green residues. D116, R123 and R127 take part to both
kinds of contacts. CBP80 residues interacting with residues
5−13 from CBP20 are depicted in pink. Hydrogen bonds and salt
bridges are represented as dashed lines, and the hydrophobic
contacts as dashed bars. (C and D) Two views of CBC bound to the
cap analogue m^7GpppG. The three MIF4G domains of CBP80 are
represented in pink, yellow and green for domains 1, 2 and 3,
respectively. CBP20 is depicted in red, and the cap analogue
m^7GpppG in cyan. (A), (B) and (C) were generated with Molscript
(Kraulis, 1991) and Render (Merritt and Murphy, 1994). NLS
complex. Ribbon representation of the cap-free conformation of
CBP20 showing residues 30−125 (grey). Residues involved in cap
binding are shown in yellow (already in their cap-bound
conformation) and blue (undergo a conformational change to
interact with the cap) (compare with B). Phe49 (light green)
changes conformation to help stabilize the C-terminal domain
(compare with B). Salt bridges and hydrogen bonds are indicated
by dashed lines. (B) Stabilization of the N- and C-terminal
extensions of CBP20 upon cap binding. Ribbon representation of
cap-bound CBP20 showing residues 32−125 (already ordered in
the cap-free form) in grey and the N- and C-terminal extensions
that fold upon cap binding in green and orange, respectively.
Yellow residues stabilize the folded conformation of these two
extensions through interactions with the cap. This stabilization
is reinforced by protein−protein interactions involving the
light green residues. D116, R123 and R127 take part to both
kinds of contacts. CBP80 residues interacting with residues
5−13 from CBP20 are depicted in pink. Hydrogen bonds and salt
bridges are represented as dashed lines, and the hydrophobic
contacts as dashed bars. (C and D) Two views of CBC bound to the
cap analogue m^7GpppG. The three MIF4G domains of CBP80 are
represented in pink, yellow and green for domains 1, 2 and 3,
respectively. CBP20 is depicted in red, and the cap analogue
m^7GpppG in cyan. (A), (B) and (C) were generated with Molscript
(Kraulis, 1991) and Render (Merritt and Murphy, 1994).
|
 |
|
 |
 |
|
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2002,
21,
5548-5557)
copyright 2002.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Co-Crystallization of the human nuclear cap-Binding complex with a m7gpppg cap analogue using protein engineering.
|
 |
|
Authors
|
 |
C.Mazza,
A.Segref,
I.W.Mattaj,
S.Cusack.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2002,
58,
2194-2197.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |
Figure 2.
Figure 2 Crystals of CBC and its complex with the cap analogue.
(a) CBC 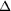 NLS,
(b) CBC NLS,
(b) CBC 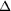 NLS NLS
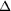 CC
with m7GpppG form 1, (c) CBC CC
with m7GpppG form 1, (c) CBC 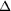 NLS NLS
 CC
with m7GpppG form 2. CC
with m7GpppG form 2.
|
 |
|
 |
 |
|
The above figure is
reproduced from the cited reference
with permission from the IUCr
|
 |
|

|
|
|
 |

